Abstract
Inheritance studies on the nonhost resistance of plants would normally require interspecific crosses that suffer from sterility and abnormal segregation. Therefore, we developed the barley–Puccinia rust model system to study, using forward genetics, the specificity, number, and diversity of genes involved in nonhost resistance. We developed two mapping populations by crossing the line SusPtrit, with exceptional susceptibility to heterologous rust species, with the immune barley cultivars Vada and Cebada Capa. These two mapping populations along with the Oregon Wolfe Barley population, which showed unexpected segregation for resistance to heterologous rusts, were phenotyped with four heterologous rust fungal species. Positions of QTL conferring nonhost resistance in the three mapping populations were compared using an integrated consensus map. The results confirmed that nonhost resistance in barley to heterologous rust species is controlled by QTL with different and overlapping specificities and by an occasional contribution of an R-gene for hypersensitivity. In each population, different sets of loci were implicated in resistance. Few genes were common between the populations, suggesting a high diversity of genes conferring nonhost resistance to heterologous pathogens. These loci were significantly associated with QTL for partial resistance to the pathogen Puccinia hordei and with defense-related genes.
PLANTS are exposed to a huge number of potential pathogens that represent a very diverse array of microorganisms. Although plants lack an immune system similar to that of mammals, susceptibility to a plant pathogen is obviously more an exception than the rule. Plants have evolved a sophisticated innate immunity network to recognize and react to potential pathogens. The genetic basis of innate immunity in plants, in particular to nonadapted (nonhost, called here “heterologous”) pathogens, is one of the most intriguing questions in the field of plant genetics, but is very hard to study since it typically requires interspecific crosses between host and nonhost species. The rare progeny that may be obtained from such crosses normally suffers from sterility and abnormal segregation, hampering the identification of individual genetic factors.
We recently developed the “Barley–Puccinia rust fungus” model system to study the inheritance and specificity of plant factors targeted by rust fungi to suppress the basal defense in barley (Jafary et al. 2006). Barley (Hordeum vulgare) is nearly a nonhost to the wheat leaf rust fungus, Puccinia triticina, and to several other heterologous rust fungal species. Few, and mainly exotic accessions of barley are somewhat susceptible to those rust species at the seedling stage (Atienza et al. 2004). In a previous study, we mapped resistance genes in a recombinant inbred line (RIL) population obtained from a cross between an immune barley line (Vada) and a research line, named SusPtrit, with exceptional susceptibility to P. triticina and to some other heterologous rust species and formae speciales (Atienza et al. 2004; Jafary et al. 2006). We showed that the nonhost resistance in Vada to heterologous rust species is controlled by sets of QTL with different and overlapping specificities and by an occasional contribution of an R-gene for hypersensitivity (Jafary et al. 2006).
Since the large majority of barley accessions is (nearly) immune to heterologous rust species (Atienza et al. 2004), one may wonder whether immunity is due to the same genes as found in Vada. To address this question, we developed an additional RIL population from a cross between the susceptible research line SusPtrit and the South American barley cultivar Cebada Capa. The latter accession is immune to heterologous rusts, like Vada and other regular barley cultivars (Atienza et al. 2004). In addition, “Cebada Capa” possesses one major gene (Rph7) for race-specific hypersensitivity resistance (HR) and several QTL for nonhypersensitive partial resistance to P. hordei (Parlevliet and Van Ommeren 1985; Qi et al. 2000). We also used a third mapping population that segregated for resistance to heterologous rust fungi, viz. the Oregon Wolfe Barley (OWB) reference mapping population (Costa et al. 2001). We aimed to map QTL involved in resistance to four heterologous rust species in Cebada Capa × SusPtrit (C × S) and OWB to compare them with the QTL identified earlier in Vada × SusPtrit (V × S) (Jafary et al. 2006). The QTL studies with the three populations allowed us to address the question of whether barley accessions commonly share resistance genes to heterologous rust species. Mapping the genes involved in nonhost resistance in the C × S and the OWB populations also verified the findings reported previously in V × S (Jafary et al. 2006) on inheritance and specificity for genes conferring nonhost resistance. Furthermore, we were able to test whether the genes involved in nonhost resistance colocate with loci for partial resistance of barley to the pathogenic barley leaf rust fungus (P. hordei) and with defense gene homolog (DGH) loci that possibly play a role in plant defense (Marcel et al. 2007).
MATERIALS AND METHODS
Plant material:
A set of RILs was obtained from a cross between the barley research line SusPtrit, which is highly susceptible to several heterologous rust fungal species in the seedling stage (Atienza et al. 2004; Jafary et al. 2006), and the immune barley cultivar Cebada Capa. From 150 F2 plants, a population of 113 F9-derived RILs was obtained by single-seed descent. The OWB (94 lines of which 2 were not available) is a doubled-haploid (DH) population derived from a cross between Dom and Rec (Costa et al. 2001). The OWB population was kindly provided by Patrick Hayes of Oregon State University.
DNA extraction and genotyping of RIL populations:
Genomic DNA of the 113 RILs and the parents SusPtrit and Cebada Capa was extracted from leaf tissue of 2-week-old seedlings according to the CTAB-based protocol of Steward and Via (1993) and adjusted for a 96-well format. The mapping population was genotyped using AFLP markers and the linkage groups were anchored to chromosomes with SSR markers. The AFLP procedure was performed according to Vos et al. (1995) with some minor modifications. The EcoRI primer with three selective nucleotides was labeled with fluorescent near-infrared labels (IRD-700 or IRD-800). AFLP fragments were separated by a denaturing polyacrylamide gel (PAGE 5.5% ready to use Gel Matrix, KB Plus, Westburg) on a LI-COR 4200 DNA automated sequencer (LI-COR Biosciences, Lincoln, NE). The scoring of the AFLP fragments was based on absence or presence of amplification products. For scoring of the AFLP fingerprints, we used the Quantar-Pro software (Keygene N.V., Wageningen, The Netherlands). In this study, 37 EcoRI/MseI primer combinations, each with three selective nucleotides, were used to generate the AFLP markers. Most of these primer combinations (25 of 37) had been used before to generate AFLP markers for construction of the L94 × Vada (Qi et al. 1998b), V × S (Jafary et al. 2006), and L94 × 116-5 maps (Qi et al. 2000). In addition, the C × S population was genotyped with a set of 14 SSR markers. The PCR product of each SSR was visualized on a LI-COR 4200 DNA automated sequencer as described for the AFLP analysis.
Marker analysis and genetic map construction:
Marker data of the C × S population were analyzed with JoinMap 3.0 (Van Ooijen and Voorrips 2001). Map distances were calculated using Kosambi's mapping function. Linkage groups were assigned to the respective barley chromosomes according to the SSR markers and AFLP markers that had already been mapped in the L94 × Vada (Qi et al. 1998b) or V × S (Jafary et al. 2006) mapping populations. The most probable order of loci within the linkage groups was estimated using the freely available (http://www.plantbreeding.wur.nl/) software RECORD (Van Os et al. 2005). A set of 242 markers was selected to construct a skeletal map for QTL mapping. Those 242 markers were homogeneously distributed and their relative positions according to JoinMap and RECORD were in agreement.
From the V × S linkage map constructed by Jafary et al. (2006), a new skeletal map was extracted with a higher density of markers, viz. 198 markers with an average distance between two consecutive markers of 5.1 cM. This skeletal map was used to repeat the QTL mapping of rust species with the data of Jafary et al. (2006).
The marker data set of the OWBs was downloaded from the barley project web site of Oregon State University (OSU) (http://barleyworld.org/). This data set was used as a component of the barley consensus map of Marcel et al. (2007). From that consensus map we extracted 206 markers mapped on the OWBs and homogeneously distributed, with an average distance between two consecutive markers of 5.1 cM, to be used as the skeletal map for QTL mapping.
Pathogen materials:
Urediospores of four heterologous rusts, P. triticina, P. persistens (old name, P. agropyrina), P. hordei–murini (Phm), and P. hordei–secalini (Phs) were multiplied on their appropriate host plants (Table 1). The spores of P. hordei isolate 1.2.1 were multiplied on susceptible barley line L98. Spores of each rust fungus were collected separately, dried for a short period (2–7 days), and stored at −80° until inoculation.
TABLE 1.
Pathogen material used in this study
| Rust species or forma specialisa | Host plant | Common name | Location of collection |
|---|---|---|---|
| P. hordei isolate 1.2.1 | Hordeum vulgare | Barley leaf rust | Wageningen, The Netherlands |
| P. hordei–murini | H. murinum | Wall barley leaf rust | Sariñena, Aragón, Spain |
| P. hordei–secalini | H. secalinum | Meadow barley leaf rust | Mesquer, Loire Atlantique, France |
| P. persistens | Elytrigia repens | Wheat grass leaf rust | Rhenen, Utrecht, The Netherlands |
| P. triticina isolate Flamingo | Triticum aestivum | Wheat leaf rust | The Netherlands |
Scientific names of rusts are used as convenient labels and do not reflect necessarily the taxonomic status of the rusts.
Phenotyping of C × S and OWB with heterologous rust fungi:
Seedlings of RILs and DHs were grown in boxes (37 × 39 cm) along with their parents and susceptible host plants. Twelve days after sowing, the first leaves were fixed in horizontal position, the adaxial side facing up. The inoculations were carried out with ∼600 spores/cm2 in a settling tower (Atienza et al. 2004). The plants were incubated overnight in a dew chamber for 10 hr (17°–18°) at 100% relative humidity and darkness and then transferred to a greenhouse compartment at ∼22°/18° (day/night). Twelve days after inoculation the level of infection was quantified by two parameters: visible infection sites per square centimeter (VIS) (the number of both flecks and pustules per square centimeter) and infection frequency (IF) (the number of pustules per square centimeter) (Jafary et al. 2006). The disease test was performed in three consecutive experiments, on three seedlings per DH line per experiment. The quantitative data for each experiment and the average of the three experiments were used as phenotypic values for QTL mapping. Resistance in OWB to Phs and to P. persistens was evaluated in two experiments, each on three seedlings per DH line.
Phenotyping C × S with P. hordei:
The RILs were grown as described above and tested for their level of partial resistance to P. hordei (isolate 1.2.1), as described by Qi et al. (1998a). The inoculum dose was ∼200 spores/cm2. The latency period (LP) of each plant was evaluated by estimating the period (in hours) at which 50% of the ultimate number of pustules became visible. The relative latency period (RLP) of seedlings was then calculated relative to the LP on seedlings of the very susceptible line L94, where LP on L94 was set at 100, as described by Parlevliet (1975). The test for RLP was performed in three consecutive experiments, on three seedlings per RIL per experiment.
For OWB, we used the phenotyping data for partial resistance as obtained by Marcel et al. (2007), but remapped the QTL.
QTL mapping:
QTL analyses in both RIL and DH populations were performed by using the MapQTL 5 software (Van Ooijen 2004). Interval mapping was performed for both IF and VIS traits, for all individual experiments and for the average over the experiments, separately. The QTL mapping procedure was followed by automatic cofactor selection, multiple-QTL mapping (MQM), and restricted MQM (Jansen and Stam 1994; Van Ooijen 1999). In the three populations, a LOD threshold of 3 was set for declaring a QTL. For significant QTL LOD-1 and LOD-2 confidence intervals were determined.
In a previous study, Qi et al. (2000) mapped the QTL for partial resistance to P. hordei present in Cebada Capa, using the L94 × 116-5 mapping population. The line 116-5 has been derived from a cross between L94 and Cebada Capa. In the F2, selection against hypersensitive resistance was performed to eliminate the Rph7 gene contributed by Cebada Capa. The remaining seedlings and their progeny from selfing were selected for long LP, to select for the genes contributing to partial resistance of Cebada Capa (Parlevliet and Kuiper 1985; Parlevliet et al. 1985). The location of the genes of Cebada Capa for partial resistance to P. hordei reported by Qi et al. (2000) were compared to those mapped in this study.
Comparison of QTL positions using an integrated consensus map:
Using an integrated map of barley (Marcel et al. 2007), we compared the distribution of QTL for nonhost resistance to find possible overlapping QTL, mapped in three different populations. This integrated map has been constructed on the basis of six mapping populations, three of which were common to this study (OWB and C × S) and previous study (V × S; Jafary et al. 2006). The map data are publicly available at the GrainGenes website (http://wheat.pw.usda.gov).
The integrated map contains 3258 markers, spanning 1081 cM with an average distance between two adjacent loci of 0.33 cM and the map has been divided into 210 bins of ∼5 cM each. In addition, 63 DGHs have been located in the bins (Marcel et al. 2007). Marcel et al. (2007) further placed 19 QTL contributing to partial resistance of barley to the pathogen P. hordei on their integrated map. Therefore, placement of the QTL for nonhost resistance identified in the three populations on the same integrated map allowed us (1) to compare the QTL positions between the three populations, (2) to find a possible association between nonhost resistance to heterologous rusts and partial resistance to P. hordei, and (3) to find a possible association between the QTL for nonhost resistance and DGHs.
Coincidence of QTL for nonhost resistance and loci for partial resistance to P. hordei on the integrated map was analyzed by counting the bins that were occupied by QTL for either trait. The bins encompassing the peak marker of a QTL were considered as occupied bins. A chi-square test was applied to test the null hypothesis, assuming independent distribution of bins occupied with a QTL for nonhost resistance and bins with a QTL for partial resistance. In the same way, we determined whether loci for quantitative resistance to heterologous rust fungi tended to coincide with molecular marker(s) corresponding to a DGH(s).
We also compiled data on QTL in barley for plant length, days to heading, and yield from the GrainGenes 2.0 databank (http://wheat.pw.usda.gov/). As many QTL as possible were assigned to a bin of the barley consensus map (Marcel et al. 2007) according to the position of their peak marker and closely linked markers. Those traits are considered unlikely to be causally associated with nonhost or partial resistance to rusts or with genes that are involved in plant defense. These data therefore served as a negative control for finding associations.
RESULTS
Construction of the genetic map:
The genetic map of C × S contained 437 markers (AFLP, SSR, and one morphological marker). On the basis of primer combination and position, we identified 156 AFLP markers in common between our map and the maps of Qi et al. (1998b) and Jafary et al. (2006). Also, the order and relative positions of 12 SSR markers were in agreement with previously published maps (Liu et al. 1996; Qi et al. 1998b; Ramsay et al. 2000; Jafary et al. 2006). The extracted skeletal map for QTL analysis comprised 242 markers, spanning 1132 cM with an average interval between two adjacent markers of 4.7 cM and a linkage group length varying between 199 cM for chromosome 2H and 127 cM for chromosome 4H. The skeletal map contained 12 intervals >10 cM, and the greatest interval was 20.6 cM, on chromosome 1HS. The complete data set of the mapping population has been deposited in GrainGenes (Barley, Cebada Capa × SusPtrit).
Phenotyping and QTL mapping in the C × S population:
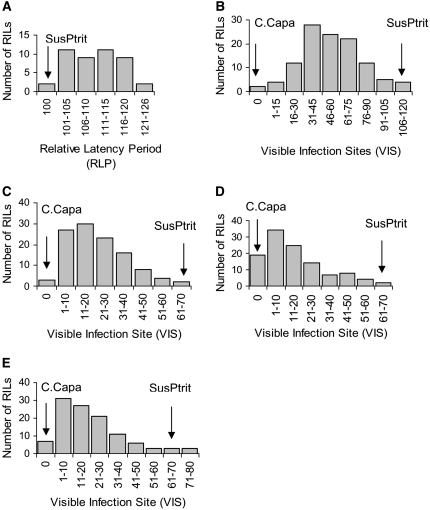
The mapping population C × S showed a quantitative segregation for the level of resistance to P. triticina, P. persistens, Phm, and Phs (Figure 1, B–E). In none of these cases was the resistance based on a macroscopically visible hypersensitive reaction. Visible infection sites that failed to develop into sporulating pustules were pale green as immature colonies. Cebada Capa was almost immune and SusPtrit was the most susceptible accession to the four rusts. The continuous and quantitative segregation for the level of infection by the four heterologous rust fungi indicates polygenic inheritance for resistance.
Figure 1.—
Frequency distribution of phenotypes for resistance to five rust species in barley mapping population Cebada Capa × SusPtrit. Values of the two parental lines are shown by arrows. For Puccinia hordei, the RLP of Cebada Capa was not measurable due to the hypersensitivity reaction. (A) P. hordei. (B) P. triticina. (C) P. persistens. (D) P. hordei–murini. (E) P. hordei–secalini.
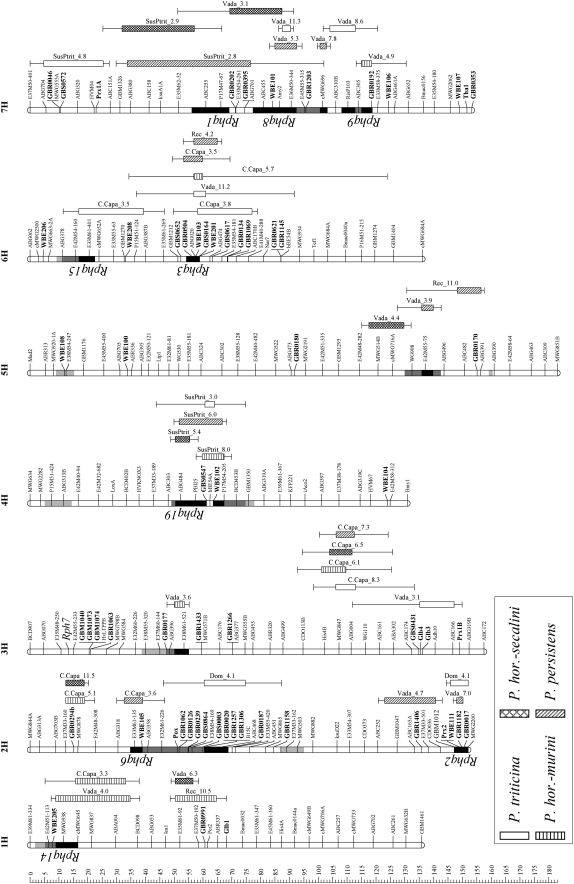
For each heterologous rust, IF and VIS gave very similar mapping results; although for three rusts (Phm, Phs, and P. triticina) VIS resulted in one additional “minor” QTL. The data presented in this report are the average VIS over the experiments of each rust fungus, and the QTL were mapped on the basis of those averages. Repeated detection of most QTL in different experiments indicates a high reliability of QTL mapping. Heritability of VIS ranged from 0.72 to 0.86, depending on the rust species. To each heterologous rust species, four or five QTL were identified (supplemental Table 1, Figure 2). The total number of chromosome regions associated with resistance to one or more rusts was nine (Figure 2). Surprisingly, for three of these regions, one on 4H and two on 7H, respectively, the resistance allele was contributed by the susceptible parent SusPtrit. Four of the nine QTL were effective to more than one rust species, i.e., had overlapping confidence intervals (Figure 2). One QTL on chromosome 3H was effective to all four heterologous rust fungi with three of them sharing the same peak marker (Figure 2, supplemental Table 1). Support intervals of another resistance QTL, contributed by the susceptible parent SusPtrit on chromosome 4H, overlapped for those rusts. There was very strong and positive correlation across the RILs between infection levels by Phs and those by Phm. For VIS, the correlation coefficient between both rusts was r = 0.92, and for IF r = 0.82, which is consistent with the finding of three chromosomal regions (2H, 3H, and, not completely overlapping, 4H) that affected the infection by both rusts Phs and Phm (Figure 2). These data suggest genes with dual effectiveness to both rusts or a close linkage of genes with a rust species-specific effect.
Figure 2.—
Locations of QTL for nonhost immunity to four heterologous rust species on a bin map extracted from a high-density consensus map of barley (Marcel et al. 2007). The QTL were originally mapped in three individual barley linkage maps. Lengths of QTL boxes (with patterns) correspond to the LOD-1 support intervals (from the peak marker) and QTL lines are extended to the LOD-2 support intervals, on the basis of results of restricted (r)MQM. The parental line contributing the allele for resistance and the LOD value obtained by rMQM are indicated on the right side of the QTL. Within chromosome bars, LOD-2 support intervals of QTL for partial resistance to barley leaf rust (Marcel et al. 2007; this study) are indicated as solid bars if overlapping with the LOD-1 support interval, as bars with dark shading if overlapping with the LOD-2 support interval, and as bars with light shading if not overlapping with QTL for nonhost resistance presented in this study. The name of the QTL for partial resistance (Rphq−) is indicated on the left side of the chromosome bars when its peak marker(s) was within the LOD-1 support interval of QTL for nonhost resistance. The 63 loci in boldface type are DGH-based markers. The ruler on the left indicates the distance in centimorgans (according to Kosambi) from the top of each chromosome.
For P. hordei, 61% of the RILs showed a hypersensitive reaction due to segregation of the Rph7 resistance allele of Cebada Capa. The segregation ratio for the HR phenotype deviated significantly from the expected 1:1 ratio (P = 0.05). However, the segregation of marker alleles in that region deviated accordingly from 1:1, indicating skewed segregation in that region. The remaining 44 RILs varied quantitatively in level of partial resistance (PR) to P. hordei, none of them causing an LP shorter than found on SusPtrit. This indicates that only Cebada Capa contributed alleles for resistance (Figure 1A). This low number of RILs was insufficient to reliably map the QTL for partial resistance to P. hordei. Qi et al. (2000) mapped three Cebada Capa-derived QTL for partial resistance in L94 × 116-5, of which one, Rphq11, with LOD score 12.0 on chromosome 2H, was confirmed at LOD 6.6 in the present C × S population (supplemental Table 1).
Phenotyping and QTL mapping in the OWB population:
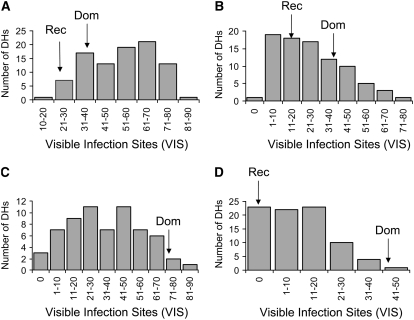
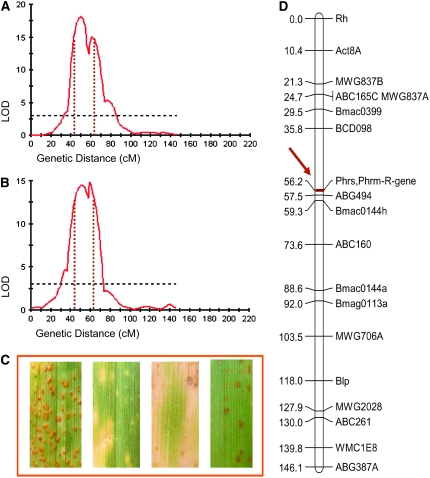
Resistance measured as IF (data not shown) and VIS to P. triticina and P. persistens (Figure 3, A and D) showed a continuous and quantitative segregation in the OWB population, without macroscopically visible HR. Heritability of the VIS ranged from 0.60 to 0.93, depending on the rust species. The two parents had a similar intermediate-resistance level to P. triticina, but differed strongly for resistance to P. persistens, Rec being immune and Dom being fully susceptible (Figure 3, A and D). This indicates a rust-specific effect of the underlying genes. Segregation for resistance to Phs and Phm was both quantitative and qualitative. Fifty-two (57%) of the DHs exhibited HR to Phm and Phs. This reaction showed as chlorotic spots, extensive chlorosis, or small, brown necrotic flecks with occasionally small pustules (Figure 4C). All DHs showing HR to Phm also showed HR to Phs. The major gene for HR reaction originated from the parent Rec and was mapped by JoinMap to the short arm of chromosome 1H (56 cM) (Figure 4D). The position of this gene is similar to that of an R-gene in the V × S population, conferring HR to Phs (Jafary et al. 2006). These data suggest the involvement of one R-gene or of an R-gene cluster that is effective to both heterologous rust fungi. The 40 DHs without the hypersensitivity gene showed various levels of infection (0–78 pustules/cm2) to both rust pathogens, indicating a segregation for additional genes with quantitative effects. The quantitative resistance to Phm and Phs could only be quantified in those 40 DH lines that did not carry the R-gene for hypersensitivity. QTL mapping for IF indicated, not surprisingly, one large-effect QTL with LOD values 18 and 15 for Phs and Phm, respectively, on the position of the R-gene for hypersensitive resistance (Figure 4, A and B) and one additional QTL that was either specific to Phs (chromosome 4H) or specific to Phm (chromosome 5H) (data not shown). QTL mapping of VIS in the lines without the hypersensitivity indicated one QTL for resistance to Phm and none for resistance to Phs (supplemental Table 1; Figure 2). The low number of lines was probably insufficient to detect all segregating QTL. In this population, a total of five QTL were detected to contribute to the VIS component of nonhost resistance of which one region corresponded to the position of the R-gene on chromosome 1H. The remaining four were effective to only one rust species.
Figure 3.—
Frequency distribution of phenotypes for resistance, measured as number of visible infection sites (VIS) per square centimeter leaf area, to four heterologous rust species in the OWB mapping population. Values of the two parental lines are shown by arrows. For P. hordei–secalini no value could be determined for parent Rec and 30 lines, because the flecks tended to merge. (A) P. triticina. (B) P. hordei–murini. (C) P. hordei–secalini. (D) P. persistens.
Figure 4.—
Mapping of an R-gene for hypersensitive resistance (HR) to Puccinia hordei–secalini and P. hordei–murini in the OWB mapping population. (A and B) LOD profiles of QTL (based on interval mapping based on number of rust pustules per square centimeter leaf area) on chromosome 1H with the highest effect to P. hordei–secalini (A) and P. hordei–murini (B). Horizontal and vertical dotted lines show LOD threshold values and the support interval of the QTL, respectively. (C) Four examples of reaction of OWB doubled-haploid lines to infection by P. hordei–secalini. From left to right: susceptible reaction, chlorotic spots, extensive chlorosis, and small necrotic flecks. (D) Position of R-gene conferring HR resistance to both P. hordei–secalini and P. hordei–murini, mapped using JoinMap software on chromosome 1H (56.2 cM) in the support interval of the QTL with the highest effect.
QTL for resistance in the V × S population:
We used earlier phenotypic data on the V × S population obtained by Jafary et al. (2006) to remap the QTL for the four heterologous rusts and for P. hordei and transferred them to the consensus map of Marcel et al. (2007) (supplemental Table 1; Figure 2). For each rust we found three to five QTL explaining the genetic variation. For all of them Vada contributed the resistance allele. There were minor differences in mapping compared with the report by Jafary et al. (2006), due to using VIS for all rusts, a skeleton map with more markers, a slightly different choice of cofactors, and a sometimes different criterion to define a QTL (see next section).
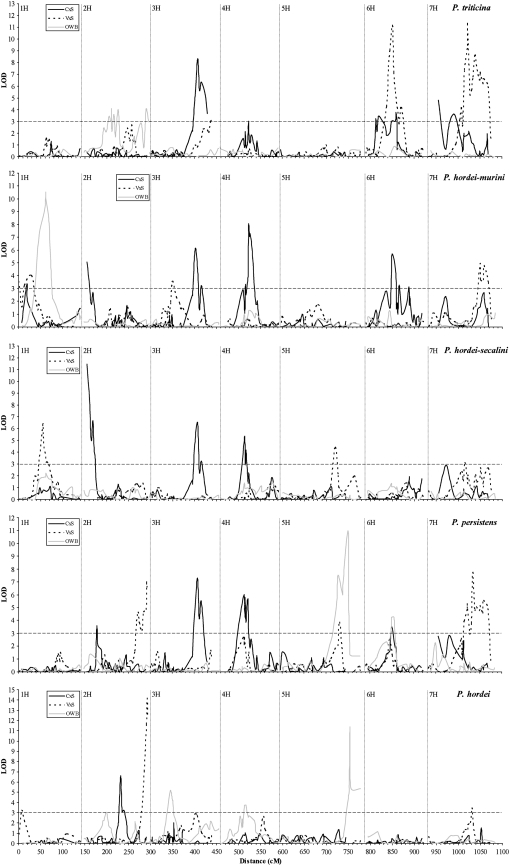
Comparison of QTL for resistance mapped in three segregating populations:
Declaring QTL and determining whether QTL overlap is often not straightforward. First, from the LOD profiles obtained, it is rather subjective which and how many QTL to consider real. The graphical representation of the LOD profiles (Figure 5) illustrates that some chromosome areas showed multiple peaks (e.g., in OWB with P. triticina on the central section of 2H and in V × S with P. persistens and P. triticina on 7H). Many clear LOD profile peaks were flanked by a second, minor peak close by (e.g., the peak on 3H of C × S for the four heterologous rusts). These phenomena may indicate two or more QTL in linkage. However, we decided to interpret the data more conservatively. When the LOD-1 confidence intervals of different peaks overlapped, we took the LOD-1 confidence interval of the highest peak as the place of the QTL and calculated the LOD-2 confidence interval as the combination of all separate LOD-2 intervals of the neighboring peaks. In two cases, the MapQTL software interpolated a LOD score surpassing LOD = 3 in the interval between linked markers, but values slightly lower than this threshold for the two flanking markers. Both QTL were contributed by SusPtrit and located on 7H (LOD 2.9 to Phs and 2.8 to P. persistens, respectively). Since they seemed to confirm each other, they were considered real (Figures 2 and 5).
Figure 5.—
LOD profiles of four heterologous rust species and of the leaf rust of barley (P. hordei) obtained by restricted multiple-QTL mapping (rMQM) in three individual barley segregating populations and plotted along a consensus map of barley (Marcel et al. 2007). The barley populations are Cebada Capa × SusPtrit (C × S), Vada × SusPtrit (S × V), and the Oregon Wolfe Barleys (OWB). Dotted vertical lines separate the seven barley chromosomes. The dashed horizontal line indicates the LOD threshold for significance.
Another complication is that QTL for resistance to the four heterologous rust species and to P. hordei in C × S and V × S populations were mapped in their respective biparental linkage map. To make direct comparison of the QTL positions possible, the positions of QTL on the respective biparental linkage map were converted to the positions on the integrated map of Marcel et al. (2007) (supplemental Table 1; Figure 2). We considered QTL regions to different rusts and/or in different populations as the same if their LOD-1 confidence intervals overlapped on the integrated map, in some cases (e.g., on 4H) in a tiling fashion. This indicated that 22 chromosomal regions were involved in resistance to at least one heterologous rust species (Figure 2). The vast majority (18) mapped in only one of the three populations. Of the 4 that mapped in two or three populations, 3 were not effective to the same rust (Figure 2). QTL to a certain rust species detected in one, but not in other populations, rarely showed a LOD peak that exceeded a LOD score of 1 in the other populations (Figure 5). This suggests that the resistance to heterologous rusts of Cebada Capa, Vada, and the OWB population is due to surprisingly different sets of genes.
Association of genes for resistance to heterologous rusts with genes for resistance to P. hordei:
The possible association between nonhost resistance and two types of host resistance (hypersensitive and partial resistance) was investigated in this study. The major gene for HR reaction to P. hordei (Rph7) of Cebada Capa was mapped by JoinMap near the top of chromosome 3H (13.4 cM on the consensus map; Figure 2). None of the QTL for resistance to heterologous rust species coincided with the position of Rph7. This indicates that Rph7 for HR resistance to P. hordei does not affect infection by heterologous rusts. In the consensus map of Marcel et al. (2007), QTL for partial resistance were indicated as they were mapped in L94 × Vada (Qi et al. 1998a), OWB and Steptoe × Morex (Marcel et al. 2007), V × S (Jafary et al. 2006), and C × S (this work). For V × S, C × S, and OWB, the QTL for resistance to P. hordei were remapped for this study, resulting in a total of 17 QTL in the five populations. The consensus map is divided into bins of ∼5 cM. The numbers of bins harboring a peak marker of a QTL for resistance to at least one of the four heterologous rusts mapped in V × S, C × S, and OWB and a peak marker for a QTL for partial resistance to P. hordei were counted to test for possible association between nonhost resistance and partial resistance (Table 2). Four QTL for partial resistance were mapped in several populations and had a different peak marker in each population. Therefore, the 17 QTL occupied 22 bins. Since R-genes are likely to govern a defense that is superimposed on the nonhypersensitive basal resistance, we did not expect a particular association between the R-gene for nonhost resistance on chromosome 1H (61.1 cM on the consensus map) of OWB with the loci for partial resistance on the consensus map. We therefore excluded that locus from our calculation. The chi-square test indicated a significant association between QTL for heterologous rusts and QTL for partial resistance, which is consistent with the hypothesis that genes for both types of resistance encode similar functions in basal defense or basic compatibility (Table 2). In total, 8 bins harbored a peak marker for nonhost resistance and a peak marker for partial resistance. Under the assumption of independent distribution of those QTL, the expected number of bins would have been 3. Of those 8 bins, in five cases resistance alleles were contributed by the same and in three cases by different barley lines. In some of the 8 bins, additional QTL mapped for which the resistance allele was contributed by a different parent. These results suggest that mostly pleiotropy and, to a lesser extent, alternative alleles explain dual effectiveness of QTL involved in resistance to host and nonhost pathogens. Since association between QTL may be due to the occurrence of the genes in gene-rich areas rather than because of functional association, we tested whether the QTL for resistance to heterologous rusts were also associated with QTL for days to heading, plant height, and yield, respectively. These traits are considered unlikely to be causally associated with nonhost or partial resistance to rusts. None of the chi-square values suggested a significant association between the QTL for resistance to heterologous rusts and the reference QTL (Table 2).
TABLE 2.
Chi-square values on the probability of independent distribution of barley QTL for partial resistance to leaf rust (QTLpr), nonhost resistance to heterologous cereal and grass rusts (QTLnh), days to heading (QTLdh), plant height (QTLph) and yield (QTLyi), and defense gene homologs (DGH), over bins on a consensus genetic map of barley
| QTLpra | QTLnh | DGH | QTLdh | QTLph | QTLyi | |
|---|---|---|---|---|---|---|
| Marker no.b | 23c | 37 | 63 | 52 | 31 | 24 |
| Bin no.d | 22 | 27 | 43 | 39 | 28 | 23 |
| QTLpr | — | 12.12e | 9.42 | 0.28 | 7.27 | 0.18 |
| QTLnh | — | — | 10.93 | 1.11 | 0.13 | 0.40 |
| DGH | — | — | — | 0.78 | 7.02 | 3.25 |
| QTLdh | — | — | — | — | 6.28 | 14.62 |
| QTLph | — | — | — | — | — | 20.31 |
Mapping data were obtained from this study (QTLpr and QTLnh), from Marcel et al. (2007) (QTLpr and DGH), and from the GrainGenes 2.0 databank (http://wheat.pw.usda.gov/) (QTLdh, QTLph, and QTLyi).
The number of (peak) markers mapped on the consensus map of barley (Marcel et al. 2007).
The chi-square values for which the null hypothesis of independent distribution is rejected with a probability P < 0.05 (with 1 d.f., P = 0.05 for χ2 = 3.841) are in italics.
The number of barley bins (5 cM) occupied by the (peak) markers for the respective class.
The chi-square values for which the null hypothesis of independent distribution is rejected with a probability P < 0.01 (with 1 d.f., P = 0.01 for χ2 = 6.634) are underlined.
Association of QTL for resistance to heterologous rusts with defense gene homologs:
The bin system of the barley integrated map was also used to test for a possible association between the distribution of loci for nonhost resistance and the distribution of 63 previously mapped DGHs (Marcel et al. 2007). The peak markers of loci for nonhost resistance occupied 27 bins, while the DGH-derived markers occupied 43 bins. We focused only on the quantitative, nonhypersensitivity class of resistance genes and therefore the R-gene for HR to Phs and Phm in OWB was excluded. Twelve bins were co-occupied by a QTL peak marker(s) and by a DGH derived marker(s). Under the assumption of independent distribution, the expected number would have been 5.5. We tested by the chi-square test the null hypothesis, assuming an independent distribution of bins occupied by a nonhost QTL peak marker(s) and bins occupied by a DGH marker(s). The null hypothesis was rejected (Table 2) with a very high probability (P < 0.001), suggesting that there is an association between the distribution of the QTL and the DGHs over the consensus map of barley. The DGH-derived markers were also significantly associated with QTL for plant height, but with a lower chi-square value (Table 2), and they were not significantly associated with the QTL for days to heading or yield. We compared the positions of the DGH-derived markers with the positions of the QTL for resistance to heterologous rusts and found that 31 of the 63 DGH-based markers mapped within the LOD-1 confidence interval of the QTL for nonhost resistance (supplemental Table 2). Ten of them were previously reported as candidate genes to explain partial resistance to barley leaf rust (Marcel et al. 2007; supplemental Table 2); the other 21 genes were not previously identified.
DISCUSSION
Near-nonhost resistance in barley:
A plant species is called host when all or the majority of accessions are susceptible to the pathogen. Full nonhost resistance can be defined as immunity, displayed by an entire plant species against all genotypes of a plant pathogen (Heath 2000). Drawing a discrete line between host and nonhost status is not always straightforward since some plant species–pathogen species combinations suggest marginal host or near-nonhost status, when only few accessions of a plant species are at most moderately susceptible to a heterologous pathogen (Niks 1987). It has been shown, for example, that barley is a full host to P. hordei, a near nonhost to some heterologous rust species, and a full nonhost to some other rust species, like P. recondita f. sp. recondita of rye (Zhang et al. 1994; Hoogkamp et al. 1998; Atienza et al. 2004). We found that wild (H. spontaneum), ancient, and exotic cultivated barleys, landraces, and accessions of barley with naked seeds have a higher chance of susceptibility to heterologous rust fungi than modern cultivars (Atienza et al. 2004). Since the parents of the OWB population have been generated by convergent crosses of exotic barley accessions (Costa et al. 2001), it was not entirely unexpected that this population would segregate for resistance to heterologous rust fungal species. Yet, understanding why some ancient cultivated and some wild barley accessions possess susceptibility genes and identifying the coevolutionary implication of carrying such genes by those accessions remain a challenge. We think that inheritance studies of near-nonhost resistance are relevant to understanding the mechanisms and genetics behind full nonhost immunity and to gaining insight into the evolutionary aspects of nonhost immunity in plants.
Diversity of loci carrying nonhost resistance:
Our data show that the complete resistance in Vada and Cebada Capa to heterologous rusts is an example of a phenotypically “qualitative” character with a quantitative inheritance.
Due to the different numbers of RILs or DHs in the mapping populations, and the fact that occasionally a major gene for resistance reduced the number of informative lines, the power to detect QTL varied per rust/population combination. Therefore, it is likely that a number of QTL, especially those with smaller effects, remained undiscovered. The majority of chromosome regions with significant LOD scores in one population lacked even small peaks in the LOD profile of other populations against the same rust species. A few significant LOD scores in one population were mirrored by low peaks in that region in another population (Figure 5), suggesting that some QTL were present in the latter population as well. Generally, however, the present data demonstrate that in each population, different sets of quantitative genes explain the resistance to heterologous rusts (Figures 2 and 5). Such a strong diversity of QTL over the three mapping populations parallels the differential expression of resistance QTL to host pathogens, reported by Bilgic et al. (2005). They found that quantitative resistance alleles in barley to spot blotch, caused by Cochliobolus sativus, appear in different map positions across three different populations, while all populations share the same resistant parent. They could rule out possible effects of epistasis between resistance loci and QTL × environment interactions, but they could not elaborate a plausible hypothesis to explain this apparent inconsistency. In our study, we presume the susceptibility alleles in SusPtrit to be the relatively rare and peculiar alleles, accumulated from various sources (Atienza et al. 2004) to result in a line with exceptional phenotype. We expected that almost all immune barleys would carry the common, i.e., resistance, alleles for almost all loci on which SusPtrit carried the susceptibility allele. Consequently, we expected almost all susceptibility alleles to appear in any cross between SusPtrit and any immune barley. The differential expression of QTL that we found instead might be due to unknown causes, as in the work of Bilgic et al. (2005). Alternatively, immune barleys may also contain some susceptibility alleles on certain QTL and therefore will not segregate for the same QTL when crossed with SusPtrit. This possibility is consistent with the observation that crossing exotic barley lines with slight susceptibility to heterologous rusts results in transgression toward increased susceptibility, leading to the research line SusPtrit with extreme susceptibility (Atienza et al. 2004). Our data suggest that, for example, Vada and Cebada Capa share only one QTL from nine for resistance to Phm and Phs (Figure 2). This suggests that crossing both immune accessions should result in at least some susceptible transgressive offspring.
Our evidence that each barley accession has a different combination of loci carrying resistance alleles to heterologous rust fungi parallels previous findings on partial resistance of barley to P. hordei. Qi et al. (2000) showed that in the crosses L94 × 116-5 and L94 × Vada, different sets of QTL for partial resistance segregated with only one of five QTL coinciding in the two populations. More recently, Marcel et al. (2007) compared the positions of QTL for partial resistance in five barley mapping populations and showed that each crossing combination segregated for different sets of QTL, with only few QTL shared by any pair of cultivars. Those data suggest that there is an abundance of loci carrying alleles for partial resistance, which is very similar to the abundance of QTL governing resistance to heterologous rust fungi.
Role of R-genes in nonhost resistance:
Our data show that the Rph7 gene for hypersensitive resistance to P. hordei carried by Cebada Capa does not confer resistance to the other rust species tested here. This confirms earlier evidence that this gene is not effective to P. triticina (Niks and Rubiales 1994; Neu et al. 2003). Niks and Rubiales (1994), however, reported that in an L94 background, the Rph7 gene conferred complete resistance to Phm (IT0 on the 0–9 scale). They used an isolate of Phm collected in the Netherlands, whereas our isolate had been collected in Aragón, Spain. Apparently, the presently used Spanish isolate is virulent to Rph7.
In the OWB population we mapped one R-gene on chromosome 1H for HR reaction to both Phs and Phm and not to P. hordei. Phs and Phm are not closely related rust fungi, the former belonging to the P. triticina group and the latter to the P. hordei group (Jafary et al. 2006). Their respective host species are both in the genus Hordeum, which may explain the dual effectiveness of this R-gene. Interestingly, in the V × S population we found one R-gene at about the same position, but that was effective to Phs and not to Phm or to P. hordei. The locus in OWB may carry an allele of the R-gene that is effective to both Phs and Phm, whereas the allele of Vada is only effective to Phs. Alternatively, the locus may be a cluster of homologous genes, of which one is effective to Phs (in Vada as well as in OWB), but another is only present in OWB and effective to Phm. Interestingly, cultivated barley, including a modern cultivar like Vada, carries an effective R-gene against a rust species (Phs) that is not pathogenic to cultivated barley and hence is very unlikely to be selected for by breeders. Our data suggest that occasionally R-genes with qualitative effects contribute to nonhost resistance in barley to heterologous rust fungi.
Specificity of QTL involved in nonhost resistance and partial resistance:
The present data (Figure 2) show that the large majority of QTL are effective to only one or two heterologous rust fungi. Only in C × S we found two QTL with overlapping confidence intervals to all four heterologous rusts. The resolution of the mapping does not allow us to conclude whether effectiveness to more than one rust fungus is due to close linkage of genes or to pleiotropy. QTL that are effective to two or more rust fungi may have such multiple effects because of the relationship between hosts (e.g., P. hordei, Phm, and Phs all infect Hordeum species) or because of the relationship between rust pathogens [e.g., P. triticina, P. persistens, and Phs are according to their ITS sequence closely related fungi (Jafary et al. 2006)]. In our data, QTL for resistance to P. hordei in one population were more often associated with QTL for nonhost resistance to P. triticina and P. persistens in the same population than to QTL for resistance to other rusts. As well, only one QTL combined effectiveness to both Phs and P. triticina, whereas five QTL were effective to P. persistens and Phs. Our data therefore do not allow preliminary conclusions whether the effectiveness of QTL depends more on the host plants than on the taxonomic position of the rust.
In all three populations, rust species specificity is the predominant characteristic of QTL contributing to nonhost resistance in barley. Such a rust species specificity of genes is further illustrated by the observation that the two parents of OWB had a similar intermediate-resistance level to P. triticina, but differed strongly for P. persistens (Figure 2, A and D). The highly significant association between QTL for partial resistance to P. hordei and QTL for resistance to heterologous rusts (Table 2) is consistent with earlier evidence that these two traits are correlated (Niks 1983a; Zhang et al. 1994; Hoogkamp et al. 1998). Indeed, both traits are based on reduced haustorium formation by the rust without substantial hypersensitivity (Niks 1983a,b). Therefore, we presume that genes governing partial resistance to P. hordei play similar roles in basal resistance as do genes for resistance to heterologous rusts.
Role of QTL genes in nonhost resistance:
Nonhost immunity is a durable and complete type of resistance in plants and may encompass passive and active defense responses (Nürnberger et al. 2004). Innate immunity includes the active defense response in plants, which is initiated by perception of pathogen-associated molecular patterns (PAMP) (Felix et al. 1999; Nürnberger et al. 2004). Perception of PAMPs by extracellular receptor-like kinases (RLK) activates PAMP-triggered immunity (PTI), which requires signals through MAP kinase cascades (Chisholm et al. 2006). Immunity of barley lines like Cebada Capa or Vada to the different heterologous rust fungi tested is not likely to be caused by preformed barriers. Immunity is more likely to be caused by the genes that are involved in compatibility or resistance, as discussed in our previous study (Jafary et al. 2006). Similar to other microbes, the rust fungi studied in this report contain PAMPs, which have a critical function in the lifestyle of the organism (Nürnberger et al. 2004). PTI therefore is probably activated in all barley accessions, whether they are immune like Cebada Capa or susceptible like SusPtrit, following the perception of PAMPs from the rust fungi. This perception should result in immunity, unless the rust fungi suppress the basal immune responses of the plant. Such a suppression of plant defense seems to be a key step for pathogenesis (De Wit 2002; Mendgen and Hahn 2002; Holub and Cooper 2004; Nomura et al. 2005). The crucial role of suppression has been demonstrated in particular for rust and powdery mildew pathogens that form long-term biotrophic relationships with their host plants and for plant pathogenic bacteria (Mellersh and Heath 2001; Panstruga 2003; Holub and Cooper 2004). This suppression is presumed to be mediated by pathogen-produced effectors. In haustorium-forming fungi, like rust and powdery mildew, haustoria may have a role in delivering such effectors into the plant apoplast (Chisholm et al. 2006). Since resistance to heterologous rusts and partial resistance to P. hordei are (predominantly) prehaustorial, we may presume a role for effectors that are delivered by the fungus into the apoplast, as has been reported for oomycete plant pathogens (Kamoun 2006), or that are delivered during the cell wall penetration stage. Some of these effectors may act as transcription factors (Lahaye and Bonas 2001); others may cleave specific cytoplasmic host proteins (Shao et al. 2003; Coaker et al. 2005). Recently, in the flax rust fungus Melampsora lini, ∼20 haustorial-secreted proteins have been identified (Catanzariti et al. 2006). Therefore, specific host genes or their products are conceivably targeted by biotrophic pathogens to achieve suppression of defense. The QTL mapped in this study are likely to be specific host factor genes, targeted by rust species to suppress basal defense. Some of the candidate genes that colocate with QTL for nonhost resistance (supplemental Table2) have been implicated in basal or nonhost resistance and therefore seem to agree with our hypothesis. For example, of the 13 colocations of QTL-peak markers with DGH markers within <1 cM we find a peroxidase gene eight times. Peroxidase genes have been implicated in basal resistance (Thordal-Christensen et al. 1997) and basic compatibility (Kristensen et al. 2001). Other interesting examples are the serine/threonine-protein kinase Pelle (HvNR-F6) gene reported by Neu et al. (2003) and the superoxide dismutase (SOD) gene. Both genes are at 0.1 and 0.2 cM, respectively, to the Cebada Capa QTL against Phm and Pper on 6H. The next challenge is to isolate some of the plant genes underlying the QTL effects to understand the plant factors that determine the success or failure of suppression of basal defense of plant cells and hence the host status of plants.
Acknowledgments
Anton Vels is gratefully acknowledged for his technical assistance. We greatly appreciate Richard Visser for reading the manuscript and giving his valuable comments and Patrick Hayes of the Oregon State University for providing seeds of the OWB mapping population. Hossein Jafary was financially supported by the Agricultural Research and Education Organisation and the Ministry of Science Research and Technology of the Islamic Republic of Iran.
References
- Atienza, S. G., H. Jafary and R. E. Niks, 2004. Accumulation of genes for susceptibility to rust fungi for which barley is nearly a nonhost results in two barley lines with extreme multiple susceptibility. Planta 220 71–79. [DOI] [PubMed] [Google Scholar]
- Bilgic, H., B. J. Steffenson and P. M. Hayes, 2005. Comprehensive genetic analyses reveal differential expression of spot blotch resistance in four populations of barley. Theor. Appl. Genet. 111 1238–1250. [DOI] [PubMed] [Google Scholar]
- Catanzariti, A. M., P. N. Dodds, G. J. Lawrence, M. A. Ayliffe and J. G. Ellis, 2006. Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell 18 243–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm, S. T., G. Coaker, B. Day and B. J. Staskawicz, 2006. Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124 803–814. [DOI] [PubMed] [Google Scholar]
- Coaker, G., A. Falick and B. Staskawicz, 2005. Activation of a phytopathogenic bacterial effector protein by a eukaryotic cyclophilin. Science 308 548–550. [DOI] [PubMed] [Google Scholar]
- Costa, J. M., A. Corey, P. M. Hayes, C. Jobet, A. Kleinhofs et al., 2001. Molecular mapping of the Oregon Wolfe Barleys: a phenotypically polymorphic doubled-haploid population. Theor. Appl. Genet. 103 415–424. [Google Scholar]
- De Wit, P. J. G. M., 2002. Plant biology—on guard. Nature 416 801–803. [DOI] [PubMed] [Google Scholar]
- Felix, G., J. D. Duran, S. Volko and T. Boller, 1999. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18 265–276. [DOI] [PubMed] [Google Scholar]
- Heath, M. C., 2000. Nonhost resistance and nonspecific plant defenses. Curr. Opin. Plant Biol. 3 315–319. [DOI] [PubMed] [Google Scholar]
- Holub, E. B., and A. Cooper, 2004. Matrix, reinvention in plants: how genetics is unveiling secrets of non-host disease resistance. Trends Plant Sci. 9 211–214. [DOI] [PubMed] [Google Scholar]
- Hoogkamp, T. J. H., W. Q. Chen and R. E. Niks, 1998. Specificity of prehaustorial resistance to Puccinia hordei and to two inappropriate rust fungi in barley. Phytopathology 88 856–861. [DOI] [PubMed] [Google Scholar]
- Jafary, H., L. Szabo and R. E. Niks, 2006. Innate nonhost immunity in barley to different heterologous rust fungi is controlled by sets of resistance genes with different and overlapping specificities. Mol. Plant-Microbe Interact. 19 1270–1279. [DOI] [PubMed] [Google Scholar]
- Jansen, R. C., and P. Stam, 1994. High-resolution of quantitative traits into multiple loci via interval mapping. Genetics 136 1447–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamoun, S., 2006. A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 44 41–60. [DOI] [PubMed] [Google Scholar]
- Kristensen, B. K., H. Ammitzboll, S. Kjærgård Rasmussen and K. A. Nielsen, 2001. Transient expression of a vacuolar peroxidase increases susceptibility of epidermal barley cells to powdery mildew. Mol. Plant Pathol. 2 311–317. [DOI] [PubMed] [Google Scholar]
- Lahaye, T., and U. Bonas, 2001. Molecular secrets of bacterial type III effector proteins. Trends Plant Sci. 6 479–485. [DOI] [PubMed] [Google Scholar]
- Liu, Z. W., R. M. Biyashev and M. A. S. Maroof, 1996. Development of simple sequence repeat DNA markers and their integration into a barley linkage map. Theor. Appl. Genet. 93 869–876. [DOI] [PubMed] [Google Scholar]
- Marcel, T. C., R. K. Varshney, M. Barbieri, H. Jafary, M. J. D. de Kock et al., 2007. A high-density consensus map of barley to compare the distribution of QTLs for partial resistance to Puccinia hordei and of defence gene homologues. Theor. Appl. Genet. 114 487–500. [DOI] [PubMed] [Google Scholar]
- Mellersh, D. G., and M. C. Heath, 2001. Plasma membrane-cell wall adhesion is required for expression of plant defense responses during fungal penetration. Plant Cell 13 413–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendgen, K., and M. Hahn, 2002. Plant infection and the establishment of fungal biotrophy. Trends Plant Sci. 7 352–356. [DOI] [PubMed] [Google Scholar]
- Neu, C., B. Keller and C. Feuillet, 2003. Cytological and molecular analysis of the Hordeum vulgare-Puccinia triticina nonhost interaction. Mol. Plant-Microbe Interact. 16 626–633. [DOI] [PubMed] [Google Scholar]
- Niks, R. E., 1983. a Comparative histology of partial resistance and the nonhost reaction to leaf rust pathogens in barley and wheat seedlings. Phytopathology 73 60–64. [Google Scholar]
- Niks, R. E., 1983. b Haustorium formation by Puccinia hordei in leaves of hypersensitive, partially resistant, and nonhost plant genotypes. Phytopathology 73 64–66. [Google Scholar]
- Niks, R. E., 1987. Nonhost plant-species as donors for resistance to pathogens with narrow host range. 1. Determination of nonhost status. Euphytica 36 841–852. [Google Scholar]
- Niks, R. E., and D. Rubiales, 1994. Avirulence factors corresponding to barley genes Pa3 and Pa7 which confer resistance against Puccinia-hordei in rust fungi other than P. hordei. Physiol. Mol. Plant Pathol. 45 321–331. [Google Scholar]
- Nomura, K., M. Melotto and S. Y. He, 2005. Suppression of host defense in compatible plant-Pseudomonas syringae interactions. Curr. Opin. Plant Biol. 8 361–368. [DOI] [PubMed] [Google Scholar]
- Nürnberger, T., F. Brunner, B. Kemmerling and L. Piater, 2004. Innate immunity in plants and animals: striking similarities and obvious differences. Immunol. Rev. 198 249–266. [DOI] [PubMed] [Google Scholar]
- Panstruga, R., 2003. Establishing compatibility between plants and obligate biotrophic pathogens. Curr. Opin. Plant Biol. 6 320–326. [DOI] [PubMed] [Google Scholar]
- Parlevliet, J. E., 1975. Partial resistance of barley to leaf rust, Puccinia-hordei. 1. Effect of cultivar and development stage on latent period. Euphytica 24 21–27. [Google Scholar]
- Parlevliet, J. E., and H. J. Kuiper, 1985. Accumulating polygenes for partial resistance in barley to barley leaf rust, Puccinia hordei. I. Selection for increased latent periods. Euphytica 34 7–13. [Google Scholar]
- Parlevliet, J. E., and A. Van Ommeren, 1985. Race-specific effects in major genic and polygenic resistance of barley to barley leaf rust in the field - identification and distinction. Euphytica 34 689–695. [Google Scholar]
- Parlevliet, J. E., M. Leijn and A. Van Ommeren, 1985. Accumulating polygenes for partial resistance in barley to barley leaf rust, Puccinia hordei. II. Field evaluation. Euphytica 29 1–8. [Google Scholar]
- Qi, X., R. E. Niks, P. Stam and P. Lindhout, 1998. a Identification of QTLs for partial resistance to leaf rust (Puccinia hordei) in barley. Theor. Appl. Genet. 96 1205–1215. [Google Scholar]
- Qi, X., P. Stam and P. Lindhout, 1998. b Use of locus-specific AFLP markers to construct a high-density molecular map in barley. Theor. Appl. Genet. 96 376–384. [DOI] [PubMed] [Google Scholar]
- Qi, X., F. Fufa, D. Sijtsma, R. E. Niks, P. Lindhout et al., 2000. The evidence for abundance of QTLs for partial resistance to Puccinia hordei on the barley genome. Mol. Breed. 6 1–9. [Google Scholar]
- Ramsay, L., M. Macaulay, S. D. Ivanissevich, K. Maclean, L. Cardle et al., 2000. A simple sequence repeat-based linkage map of barley. Genetics 156 1997–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, F., C. Golstein, J. Ade, M. Stoutemyer, J. E. Dixon et al., 2003. Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301 1230–1233. [DOI] [PubMed] [Google Scholar]
- Steward, C. N., and L. E. Via, 1993. A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques 14 748–750. [PubMed] [Google Scholar]
- Thordal-Christensen, H., Z. G. Zhang, Y. D. Wei and D. B. Collinge, 1997. Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 14 1187–1194. [Google Scholar]
- Van Ooijen, J. W., 1999. LOD significance thresholds for QTL analysis in experimental populations of diploid species. Heredity 83 613–624. [DOI] [PubMed] [Google Scholar]
- Van Ooijen, J. W., 2004. MapQTL 5, Software for the Mapping of Quantitative Trait Loci in Experimental Populations. Kyazma B.V., Wageningen, The Netherlands.
- Van Ooijen, J. W., and R. E. Voorrips, 2001. JoinMap Version 3.0, Software for the Calculation of Genetic Linkage Maps. Plant Research International, Wageningen, The Netherlands.
- Van Os, H., P. Stam, R. G. F. Visser and H. J. Van Eck, 2005. RECORD: a novel method for ordering loci on a genetic linkage map. Theor. Appl. Genet. 112 30–40. [DOI] [PubMed] [Google Scholar]
- Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. Vandelee et al., 1995. AFLP - a new technique for DNA-fingerprinting. Nucleic Acids Res. 23 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. S., R. De La Rosa, D. Rubiales, H. H. Lubbers, J. W. Molenveld et al., 1994. Role of partial resistance to Puccinia hordei in barley in the defense of barley to inappropriate rust fungi. Physiol. Mol. Plant Pathol. 45 219–228. [Google Scholar]