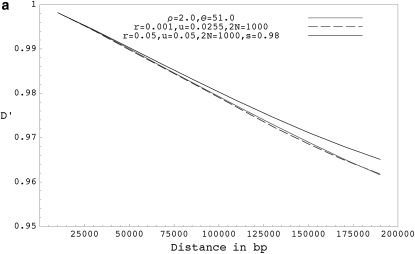
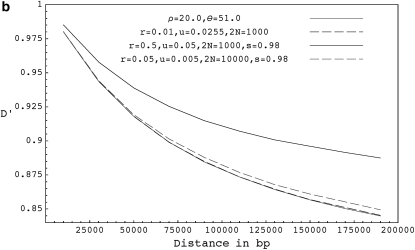
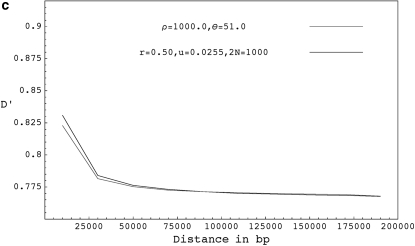
Figure 3.—
(a) The expected decay of pairwise D′ for coalescent simulations with ρ = 2.0 and θ = 51.0 (shaded curve), forward simulations with r = 0.001, u = 0.0255, and 2N = 1000 (solid dashed curve), and forward simulations with selfing for r = 0.05, u = 0.05, 2N = 1000, and s = 0.98 (solid curve). (b) The expected decay of pairwise D′ for coalescent simulations with ρ = 20.0 and θ = 51.0 (shaded curve), forward simulations with r = 0.01, u = 0.0255, and 2N = 1000 (solid dashed curve), forward simulations with r = 0.5, u = 0.05, 2N = 1000, and s = 0.98 ( solid curve), and forward simulations with r = 0.05, u = 0.005, 2N = 10,000, and s = 0.98 (shaded dashed curve). (c) The expected decay of pairwise D′ for coalescent simulations with ρ = 1000.0 and θ = 51.0 (shaded curve), and forward simulations with r = 0.50, u = 0.0255, and 2N = 1000 (solid curve). D′ values shown are based on a sample size of 20 chromosomes collected from the final populations and are averaged over 10,000 runs. Forward simulations were run for 40N generations.