Figure 1.
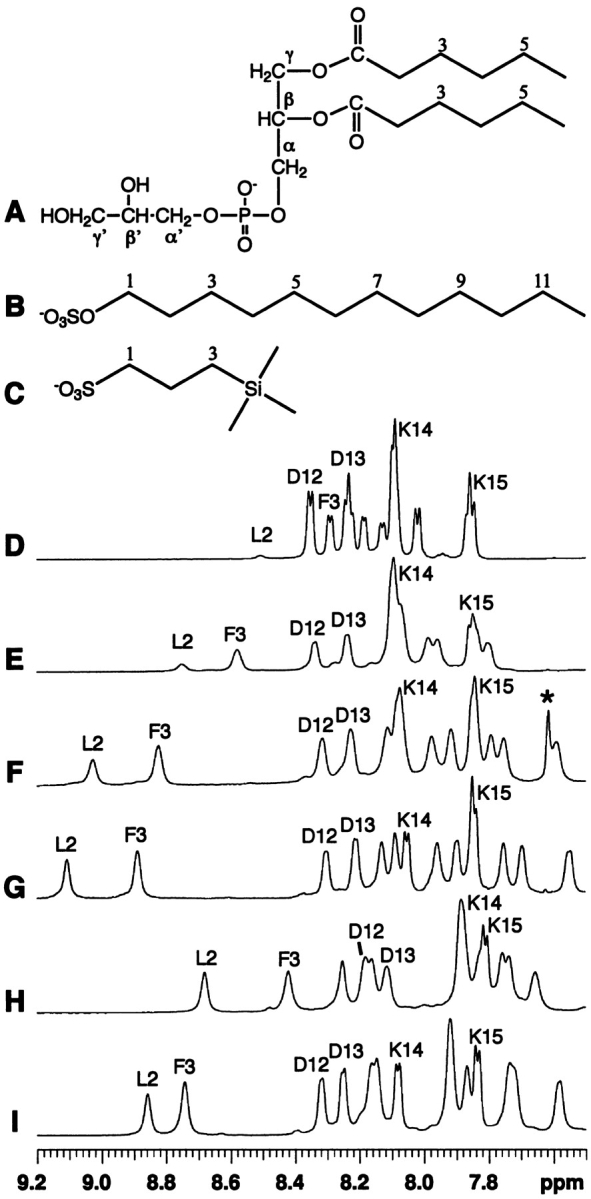
Chemical structures of detergents (A–C) and their effects on the NMR spectra (D–I) of a peptide corresponding to the N-terminal domain of enzyme IIAGlc of E. coli. Shown here are spectra in water (D), in the presence of dihexanoyl phosphatidylglycerol (DHPG, structure A) at a peptide:DHPG molar ratio of 1:2 (E), 1:5 (F), and 1:10 (G), sodium dodecylsulfate (SDS, structure B) at a peptide:SDS ratio of 1:20 (H), and 2,2-dimethyl-2-silapentane-5-sulfonate sodium salt (DSS, structure C) at a peptide:DSS ratio of 1:40 (I), at pH 5.4 and 25°C. The resonance for residual chloroform at 7.62 ppm in F was labeled by a star and was removed after evaporation under vacuum overnight (G). The peptide amide signals of Leu 2, Phe 3, Asp 12, Asp 13, Lys 14, and Lys 15 are labeled using the single-letter amino-acid code. At 25°C and pH 5.4, the chemical shifts of DHPG (A) in water are C2-H, 2.40 ppm; C3-H, 1.59 ppm; C4-H, 1.29 ppm; C5-H, 1.29 ppm; C6-H, 0.86 ppm; CH2α, 4.03 ppm; CHβ, 5.29; CH2γ, 4.27, 4.44; CH2α′, 3.89 ppm; CHβ′, 3.84 ppm; and CH2γ′, 3.59, 3.66 ppm. The chemical shifts of SDS (B) in water are C1-H, 4.02 ppm; C2-H, 1.68; C3-H, 1.36 ppm; C(4–11)-H, 1.30 ppm; and C12-H, 0.89 ppm. The chemical shifts of DSS (C) in water are C1-H, 2.90 ppm; C2-H, 1.75 ppm; C3-H, 0.63 ppm; and CH3 (triple methyls), 0.00 ppm. In the presence of the peptide, the signals of SDS and DHPG were found to shift upfield slightly by ~0.05 ppm and ~0.01–0.02 ppm, respectively, whereas the signals of DSS were found to shift downfield by ~0.01–0.02 ppm. The sodium ions are omitted to emphasize the anionic nature of these compounds.
