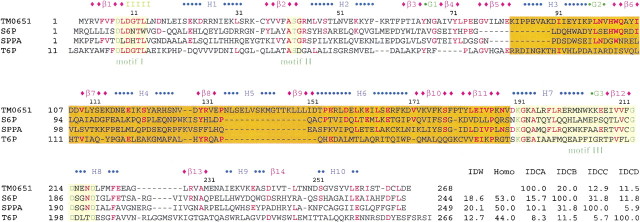
Figure 1.
Sequence comparison between TM0651 and its homologs. Several homologs of TM0651, all with defined substrate specifity, were selected for a sequence comparison. Abbreviations are as follows: (S6P) Sucrose-6F-phosphate phosphohydrolase from Synechocystis sp. PCC 6803 (Lunn et al. 2000), (SPPA) sucrose-phosphate phosphatase (sucrose-6-phosphate hydrolase) from Nostoc sp. PCC 7120 (gi:14594809; http://www.ncbi.nlm.nih.gov), (T6P) trehalose-6-phosphate phosphatase from E. coli (Giaever et al. 1988), (IDW) % whole sequence identity with TM0651, (Homo) % whole sequence homology with TM0651, (IDCA, IDCB, IDCC, IDCD) relative % cap domain sequence identities versus TM0651, S6P, SPPA, and T6P, respectively. The sequence alignment was done based on sequence comparison and the secondary structure of TM0651. The secondary structure derived from TM0651 is shown above the sequence. The blue character H represents a sequence belonging to an α-helix, a green G for 310-helix, a pink β for β-strand, and "I" for the pi-helix. The sequence identical to TM0651 is colored red. The three motifs are represented as a light yellow box with essential active site residues in dark green characters. The cap domain (residues from 82 to 189) is boxed with scarlet. The "—"s represent gaps and the residue numbers refer to those of TM0651.