Figure 2.
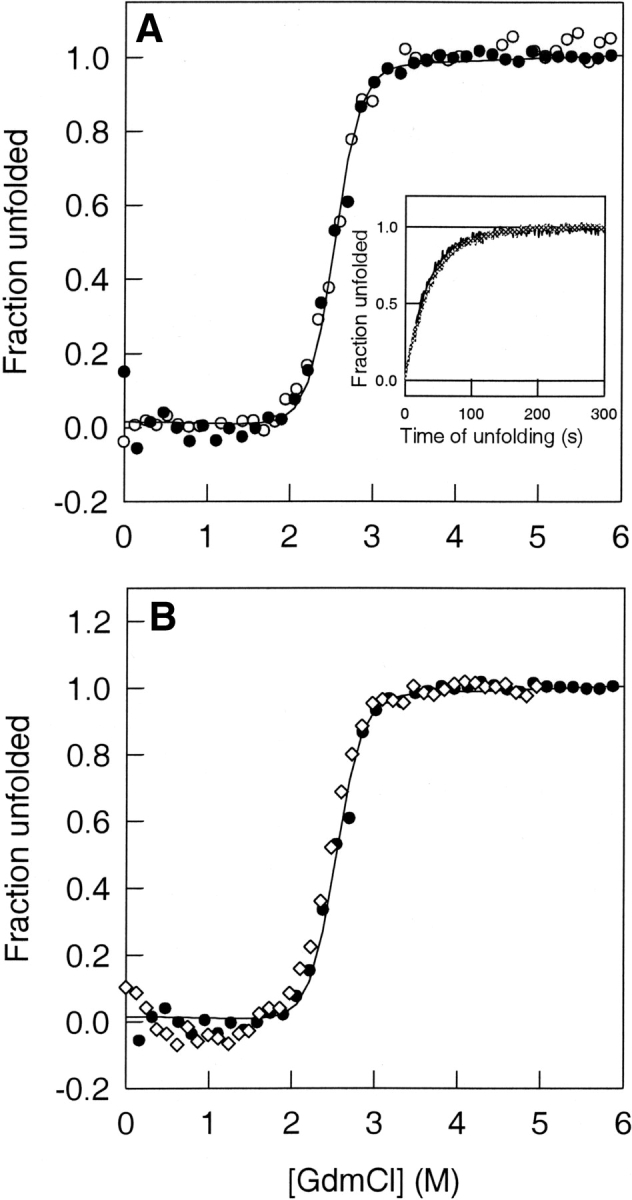
Global unfolding of thioredoxin. (A) The fraction of unfolded Trx molecules, as monitored by fluorescence (open circles) at 368 nm and CD (filled circles) at 222 nm, is plotted against the concentration of GdmCl in which Trx was equilibrated at pH 7. The solid line through the data is a fit of the data to a two-state N ⇌ U model (Agashe and Udgaonkar 1995) and yields a CM of 2.5 M. The inset to the figure shows the kinetics of unfolding of thioredoxin in 3.2 M GdmCl, followed by both fluorescence and CD. In either case, a fit (solid lines) to a single exponential yielded a rate of unfolding of 0.02 sec−1. In both cases, the data were normalized to a value of 0 for the native protein and to a value of 1 for the unfolded protein. (B) Equilibrium unfolding transitions of Trx in H2O (filled circles) and D2O (open diamonds) buffers at pH 7. The fraction of unfolded Trx molecules is plotted against the GdmCl concentration in which the protein is equilibrated. The equilibrium melts overlap, showing that there is no stabilization of Trx in D2O. The solid line through the data is a fit of the data to a two-state N ⇌ U model (Agashe and Udgaonkar 1995).
