Abstract
Markers overexpressed in colonic tumors of the multiple intestinal neoplasia (Min) mouse have been recently identified by cDNA subtractive hybridization and by microarray analysis. The significance of such a marker depends on its expression in tumor vs stromal lineages and on its expression pattern in normal tissue. From 34 differentially expressed markers, 14 were found to be expressed from supporting lineages. The markers expressed in the tumor lineage were grouped into three classes on the basis of ISH in mouse models and IHC in human adenomas. The first class includes markers expressed both in neoplastic cells and in the proliferating cells residing at the bottom of normal colonic crypts. The second class of markers shows elevated expression in neoplastic cells and also in the postmitotic Paneth cells of the small intestine. Finally, the third class of marker shows detectable intestinal expression only within tumors but not in the normal intestinal epithelium. Is such a tumor-associated marker uniquely essential for tumor growth? Deficiency for the tumor-associated glycoprotein clusterin does not affect the multiplicity or growth rate of intestinal tumors in Min mice. Thus, clusterin is a candidate secreted colon cancer marker but not a single target for chemoprevention or therapy. (J Histochem Cytochem 56:433–441, 2008)
Keywords: colorectal cancer, gene expression, Min mouse, intestinal tumor, clusterin
Colorectal cancer is one of the most common cancers in the Western world, with high morbidity and mortality (American Cancer Society 2007). Here, loss of the function of adenomatous polyposis coli (APC) protein leads to tumor formation (Powell et al. 1992; Hugh et al. 1999). In recent decades, numerous mouse models have been developed for cancer research (Bedell et al. 1997), including the multiple intestinal neoplasia (Min) mouse model of intestinal cancer (Su et al. 1992). C57BL/6(B6) mice heterozygous for this allele all develop multiple intestinal adenomas by 90 days of age (Moser et al. 1990; Dove et al. 1998). Because of its reliable phenotype, many aspects of this model have been analyzed, including the alteration of gene expression during colonic tumorigenesis (Paoni et al. 2003; Leclerc et al. 2004).
Colorectal neoplasms carry a number of mutations, some of which affect the cancer phenotype as shown by human and mouse kindreds predisposed to colon cancer (Sjoblom et al. 2006). Beyond these mutated genes, numerous others show altered levels of expression in tumor tissue compared with the normal epithelium. Because both tissues contain diverse cell types, these alterations in transcript levels may reflect changes either within a particular cell type or in the representation of different cell types. Recently, gene expression profiling for colorectal cancer has been performed using cancer cell lines (Zhang et al. 1997; Buckhaults et al. 2001), biopsy samples (Muro et al. 2003; Williams et al. 2003), or animal models for human cancers (Paoni et al. 2003; Leclerc et al. 2004). Various approaches have generated long lists of candidate genes of interest. Analysis of normal and tumor tissue at cellular resolution, combined with determining the phenotypic consequences of mutations, in particular misexpressed genes, will provide insight into the biological meaning of these candidates.
Here, we report the analysis of the cellular expression patterns of 20 candidate genes detected by suppression subtractive hybridization (SSH) and microarray analysis (MA) (Kaiser et al. 2007). Each of these shows elevated expression in the neoplastic cells of colonic adenomas isolated from ApcMin mice. The expression patterns, at a cellular level within neoplastic and normal tissues from mice and humans, were analyzed by ISH and IHC. Fourteen of the candidates that were expressed outside the tumor lineage, including hemoglobin, will be the subject of a separate study. The genes expressed in the tumor lineage can be divided into three classes according to their expression patterns in the normal intestinal epithelium. We showed that mutation of the clusterin gene, which is expressed only in tumors, does not affect any measurable aspect of tumorigenesis in the intestine of Min mice.
Materials and Methods
Mice
Mice were bred, maintained, and genotyped for the Min mutation (Su et al. 1992) in the animal facility of McArdle Laboratory, which is approved by the American Association of Laboratory Animal Care. All experiments were carried out in accordance with protocols approved by the Animal Care and Use Committee in accordance with the NIH Guide for the Care and Use of Laboratory Animals. The ApcMin mice on the C57BL/6J (B6) background were obtained from the Jackson Laboratory (Bar Harbor, ME). Mice carrying the clusterin knockout allele on a B6 background were established by B.J.A. (Fink et al. 2006).
Tissue Collection and RNA Preparation
Min mice were euthanized and dissected at 100 days of age. After isolation of the total intestinal tract, the tumors and their adjacent normal tissues were isolated individually and fixed immediately in RNAlater, RNA stabilization reagent (10 ml/g of wet weight; Qiagen, Valencia, CA). Total RNA and mRNA were isolated with TRIzol reagent (Invitrogen; Carlsbad, CA) and a Poly(A) Purist mRNA Isolation Kit (Ambion; Austin, TX), respectively, according to the manufacturers' protocols. For histology, the tumors from the small intestine, the cecum, and the colon, along with adjacent normal tissues, were isolated at defined ages, fixed immediately in 10% formalin solution for 24 hr, and stored in 70% ethanol. Fixed tissues were dehydrated and processed under RNase-free conditions to produce 5-μm paraffin sections on positively charged slides.
Human Tissues
Archived human colonic adenomas were kindly provided by Dr. Jose Torrealba (Department of Pathology and Laboratory Science, University of Wisconsin-Madison, Madison, WI).
ISH
Specific primers (20 bp) were designed for each gene of interest to generate cDNA fragments (∼0.5 kbp) with total RNA from colonic tumors using the Titan One RT-PCR system (Roche; Indianapolis, IN), according to the manufacturer's protocol. To generate the templates for ISH probes of a gene, one primer was linked to the T7 RNA polymerase promoter (5′-CTAATACGACTCACTATAGGG-3′) on its 5′ end and combined with the other primer for the PCR, in which the cDNA fragment served as the template. The T7 promoter tag was linked to the reverse primer for the antisense probe and to the forward primer for the sense control probe. The resulting DNA fragments carrying the T7 promoter were gel-purified, and used for in vitro transcription with T7 polymerase (Roche) and a digoxigenin-labeled NTP mix (Roche) to synthesize labeled cRNA probes (antisense and sense), according to the manufacturer's instructions. The synthesized probes were checked by gel electrophoresis for quality. Non-radioactive ISH was performed on the paraffin sections as described previously (Chen et al. 2003).
IHC
Antibodies against the 70-kDa heat shock protein (HSP70), matrix metalloproteinase 7 (MMP7), and MARCKS-like protein (MLP; monoclonal, Chemicon, Temecula, CA) were used (1:100 dilution) for IHC. IHC was performed on paraffin sections of mouse and human tumors with Histostain Plus [diaminobenzidine tetrahydrochloride (DAB)] kit and HistoMouse (DAB) kit (Zymed Laboratories; South San Francisco, CA), according to the manufacturer's instructions.
Intestinal Tumor Scoring and Sizing
All mice were killed by CO2 asphyxiation at 100 ± 3 days. The entire small intestine and colon were removed, opened longitudinally, cleaned, and fixed as previously described (Dietrich et al. 1993). Intestinal tumors were scored from fixed tissues under an Olympus dissecting microscope at ×10 magnification. All tumor scoring was performed by a single observer (X.C.) blind to the genotypes of the mice. For tumor sizing vs clusterin genotype, 10 representative mice were randomly selected from each of three groups. The maximum diameters of all tumors in these selected mice were measured with a calibrated eyepiece reticule in a Nikon SMN-Z stereomicroscope (Nikon; Melville, NY).
Results
Identification of Genes Whose Expression Is Elevated in Min Intestinal Tumors
A group of genes, upregulated in colonic tumor tissue in Min mice, was identified using two different approaches: expression in tumors compared with that of adjacent normal tissue by SSH and expression in tumors was compared with that during normal development using MAs (Kaiser et al. 2007). For further validation, we chose 34 candidate genes from both approaches that have markedly elevated expression in Min colonic tumors.
To study transcript levels in murine colonic tumors at a cellular level, ISH was performed with gene-specific cRNA probes and corresponding sense control probes. We chose both Min tumors and tumors induced by azoxymethane (AOM), a strong carcinogen, because both of them showed strong cytoplasmic and nuclear β-catenin accumulation (Kaiser et al. 2007). Of these 34 genes, 20 showed tumor-autonomous expression on tumor sections. The other 14 genes lacked detectable expression within the tumor lineage itself and will be studied separately. Although a positive signal for each of 20 candidate genes was observed within tumor sections, the distribution and expression levels were quite distinct. Based on their expression patterns, these genes can be divided into at least three groups. The three patterns are not mutually exclusive; they coexist in serial sections of individual colonic tumors (Table 1 ): proliferation zone associated, Paneth cell associated, and tumor associated.
Table 1.
List of genes with elevated expression within Min tumors
| Gene name | Symbol | Detection | Expression | Distribution in non-neoplastic tissues |
|---|---|---|---|---|
| Matrix metalloproteinase 7 | MMP7 | SSH | Paneth | Paneth cells of small intestine |
| Defensin α1 | DefA1 | SSH | Paneth | Neutrophils, Paneth cells of small intestine |
| Lysozyme | Lyz | SSH | Paneth | Paneth cells of small intestine |
| Heterogeneous nuclear ribonucleoprotein A1 | hnrPA1 | SSH | PZA | Nuclei of eukaryotic cells |
| Heat shock protein 70 | HSP70 | SSH | PZA | Muscle, heart, esophagus, brain, testis |
| β-Tubulin 5 | Tubb5 | SSH | PZA | Microtubules |
| Synaptophysin-like protein | Sypl | MA | PZA | Synaptic vesicle protein, pancreas |
| Stathmin 1 | Stmn1 | MA | PZA | Universal, high in mitotic cells |
| CD24 antigen | CD24 | MA | PZA | B-lineage cells, granulocyte |
| Arachidonate 12-lipoxygenase | Alox12 | MA | PZA | White blood cells |
| Protein expressed in non-metastatic cells | NME4 | SSH | PZA | Prostate, heart, liver, small intestine, and skeletal muscle |
| Clusterin | Clu | SSH | Tumor | Widely distributed |
| Tissue-type plasminogen activator | tPA | SSH | Tumor | Endothelial cells; nervous system, regions with cell migration |
| Wnt inhibitory factor 1 | WIF1 | MA | Tumor | Retina, lung, mammary gland |
| Caspase 6 | Casp6 | MA | Tumor | Apoptotic cells |
| MARCKS-like protein | MLP | MA | Tumor | Widely distributed, high in testis and uterus |
| Tescalcin | Tsc | MA | Tumor | Heart, brain, and stomach |
| SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2 | Spock2 | MA | Tumor | Brain, thymus, peripheral blood leukocyte |
| Lysosomal membrane glycoprotein 2 | LAMP2 | MA | Tumor | Placenta, lung, and liver |
| Potassium voltage-gated channel, Isk-related subfamily, gene 3 | KCNE3 | MA | Tumor | Muscle |
Proliferation Zone–associated Expression
The HSP70 protects cells from stress by binding and stabilizing partially folded proteins (Wegele et al. 2004). ISH showed strongly elevated expression of HSP70 within Min colonic tumors (Figures 1A and 1B), with both strong and weak staining in same tumor. Strong expression was also observed at the base of normal crypts, which contains proliferating cells derived from the intestinal stem cells. Similar expression of HSP70 was detected in the adenomas and bottom of crypts of the small intestine (Figure 1C). In addition, the same expression pattern of HSP70 was observed in colonic adenomas induced by AOM (Figure 1D).
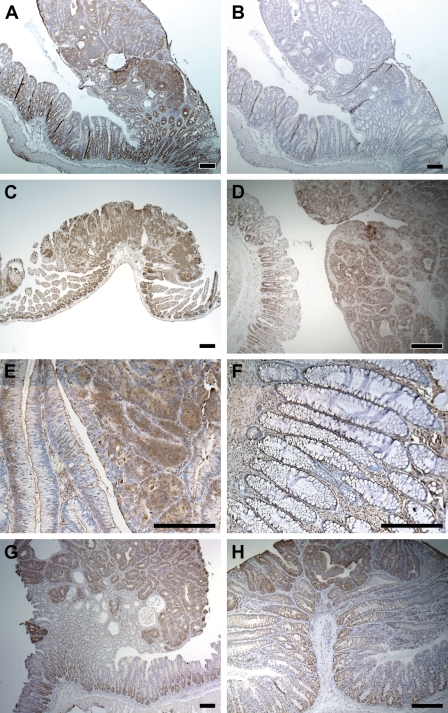
Figure 1.
Proliferation zone–associated expression. (A) Strong staining of the 70-kDa heat shock protein (HSP70) RNA in a multiple intestinal neoplasia (Min) colonic adenoma and in the proliferation zone of adjacent normal crypts. (B) Negative control of A with sense probe. (C) Elevated staining of HSP70 RNA in a Min adenoma from the small intestine and in the proliferation zone of adjacent normal crypts. (D) Strong staining of HSP70 in a colonic tumor induced by azoxymethane (AOM). (E) Immunostaining showing strong production of HSP70 protein in a human colonic adenoma. (F) Apparent IHC signal of HSP70 protein in the lower part of normal epithelial crypts. (G) Strong RNA signal of stathmin1 in a Min colonic tumor. (H) Strong signal of CD24a in a Min colonic tumor indicated by ISH. Bar = 200 μm.
IHC of human colonic adenomas showed strong staining of HSP70 in both adenomas (Figure 1E) and adenocarcinomas (data not shown), as well as in the bottom of the normal crypts (Figure 1F). Therefore, HSP70 expression in human colonic lesions qualitatively matches that in the murine model.
Several other genes showed this proliferation zone–associated expression pattern: stathmin 1 (Figure 1G), CD24a antigen (Figure 1H), heterogeneous nuclear ribonucleoprotein A1 (hnRPA1), β-tubulin 5 (tubb5), synaptophysin-like protein, arachidonate 12-lipoxygenase, and protein expressed in non-metastatic cells (NME4; data not shown for the last five genes). Immunostaining for stathmin 1 and CD24a on human samples showed similar expression in the proliferation zone of normal colonic crypts (data not shown).
To compare this expression pattern with that of Ki67, a proliferation marker, ISH for CD24a antigen and IHC for Ki67 were performed on adjacent sections of a Min colonic tumor. Interestingly, the comparison between these two showed no apparent correspondence within the tumors, indicating that genes with proliferation zone expression in normal tissue may function differently in tumors or be passive recruits.
Paneth Cell–associated Expression
MMP7, or matrilysin, degrades extracellular matrix, and therefore is involved in normal and pathogenic tissue remodeling (Ii et al. 2006). MMP7 is present in the granules within Paneth cells (Wilson et al. 1999), located at the bottom of crypts in the small intestine. Strong punctate expression of MMP7 was detected within colonic adenomas but was absent from the normal colonic epithelium (Figure 2A ). Similar staining can be detected in Min adenomas from the small intestine (Figure 2C) and at the bottom of normal crypts in the small intestine, where Paneth cells reside (Figure 2C). Punctate expression of MMP7 was also detected in the colonic tumors induced by AOM (Figure 2D).
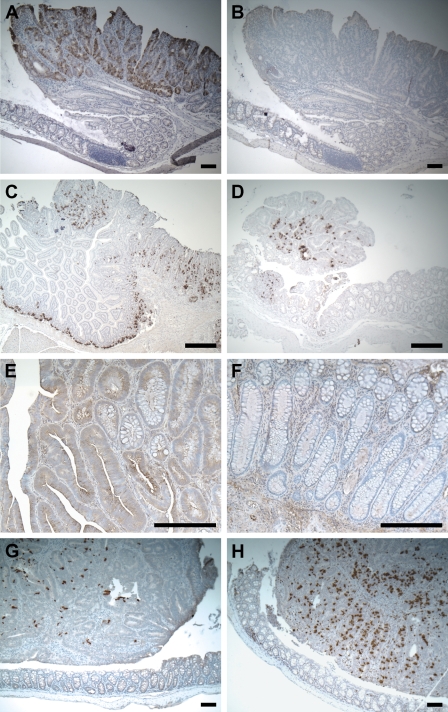
Figure 2.
Paneth cell–associated expression. (A) Strong punctate ISH signal of matrix metalloproteinase 7 (MMP7) RNA in a Min colonic adenoma. (B) Negative control of A with sense probe. (C) Punctate ISH signal of MMP7 RNA in a Min adenoma from small intestine and in Paneth cells at the bottom of adjacent normal crypts. (D) Strong ISH signal of MMP7 in a colonic tumor induced by AOM. (E) Immunostaining showing strong production of MMP7 protein in a human colonic villous adenoma. (F) No MMP7 antigen detected in the normal human colonic epithelium. (G) Punctate ISH signal of defensin α1 RNA within a colonic adenoma. (H) Strong punctate ISH signal of lysozyme M RNA within a Min colonic adenoma. Bar = 200 μm.
A polyclonal antibody against MMP7 antigen was used to detect MMP7 expression in human colonic neoplasms. Strong positive signals were detected within adenomas (Figure 2E) and adenocarcinomas (data not shown) but not in normal colonic epithelium or in hyperplastic polyps (Figure 2F), which seem to be an entity distinct from the adenoma. This result is consistent with our observations in the murine model. Immunostaining for lysozyme showed strong staining on human colonic adenomas (data not shown).
Two other genes, defensin α1 (Figure 2G), and lysozyme M (Figure 2H), shared a similar expression pattern within normal and neoplastic intestinal epithelium: strong expression within the Paneth cells of the small intestine and strong punctate staining within Min tumors arising in the colon. Because all of these genes have been traditionally recognized as markers for intestinal Paneth cell markers, these three genes were grouped together and represent a unique pattern: Paneth cell–associated expression. Recently, van Es et al. (2005) have reported that Paneth cell–associated genes can be induced by aberrant Wnt signaling in intestinal neoplasia.
Tumor-associated Expression
MLP or MacMARCKS is a well-characterized protein kinase C substrate (Lobach et al. 1993) whose expression can be strongly induced in macrophages by bacterial lipopolysaccharide (Li and Aderem 1992). In addition, this gene is critical for early neural development (Chen et al. 1996). MA identified MLP as a candidate gene strongly upregulated within Min colonic tumors. ISH with MLP-specific probes detected strong positive signals within Min colonic adenomas but not in the adjacent normal epithelium (Figure 3A ). A similar pattern was also detected in adenomas from the small intestine (Figure 3C). Unlike the punctate expression of MMP7, the staining of MLP mRNA is uniform among adjacent cells. Different regions of the tumor, however, may have quite different expression levels, resulting in “patchy” staining. Similar patterns were observed on tumors induced by AOM (Figure 3D) but not in adjacent normal epithelium (Figure 3D, inset).
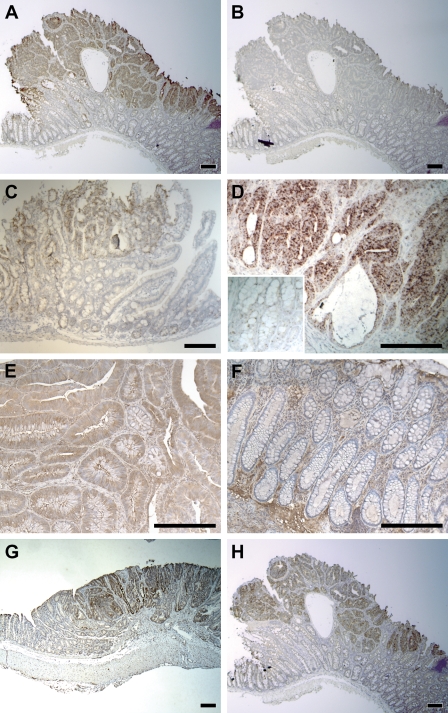
Figure 3.
Tumor-associated expression. (A) Strong ISH signal of MARCKS-like protein (MLP) RNA in a Min colonic adenoma but not in the adjacent normal crypts. (B) Negative control of A with sense probe. (C) Elevated ISH signal of MLP RNA in a Min adenoma from the small intestine. (D) Elevated ISH signal of MLP RNA in a colonic tumor induced by AOM (inset: adjacent normal epithelium at the same magnification). (E) Immunostaining showing strong production of MLP antigen in a human colonic adenoma. (F) Absence of MLP antigen in the normal human colonic epithelium. (G) Tumor-associated ISH signal of clusterin RNA in a Min colonic adenoma. (H) Strong ISH signal of Wnt-inhibitory factor 1 (WIF-1) RNA within a Min colonic adenoma but not in the adjacent normal epithelium. Bar = 200 μm.
A monoclonal antibody against human MLP was used to detect MLP antigen on human colonic lesions. Elevated MLP production can be detected in a subset of tumor cells of the colonic tubular (data not shown) and villous adenomas (Figure 3E) but not in adjacent normal epithelium (Figure 3F) or hyperplastic polyps. Similar staining can be found within invasive human adenocarcinomas (data not shown).
Unlike the patterns associated with the normal proliferative zone and with the Paneth cell, elevated MLP expression can be detected only in neoplastic tissue and not in normal intestinal tissue. Immunostaining for β-catenin on adjacent slides indicated that MLP staining lay within tumor cells as defined by strong accumulation of β-catenin caused by loss of wildtype APC (data not shown). This expression pattern was therefore defined as tumor associated. We identified 10 other genes with tumor-associated expression pattern: clusterin (Chen et al. 2003) (Figure 3G); Wnt-inhibitory factor 1 (WIF-1) (Figure 3H); lysosomal membrane glycoprotein 2 (LAMP2); tissue-type plasminogen activator (tPA); caspase 6; tescalcin; SPARC/osteonectin, CWCV, and Kazal-like domains proteoglycan 2 (Spock2); and potassium voltage-gated channel, Isk-related subfamily, gene 3 (Kcne3) (data not shown for the last six genes). Depending on the availability of suitable antibodies, IHC could be performed on human adenomas for clusterin, WIF-1, LAMP2, and tPA. All four genes showed tumor-associated expression in human samples.
Testing an Effect of Clusterin Deficiency on the Tumor Phenotype of Min Mice
A gene recruited for expression in intestinal tumors and not expressed in normal intestinal tissue might have an essential function in the biology of that tumor. In this case, ablation of such a gene would affect tumorigenesis. Clusterin is a candidate gene with confirmed tumor-associated expression.
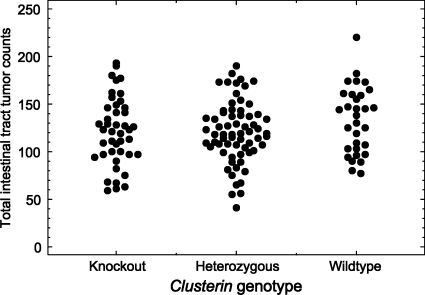
To study whether clusterin deficiency affects Min-induced intestinal tumorigenesis, Min mice were bred to those carrying the knockout allele of clusterin, and the resulting progeny were intercrossed. At 100 days, Min mice carrying zero, one, or two copies of the clusterin-deficient allele were killed, and their entire intestinal tracts were isolated and fixed. The average tumor multiplicity for the three genotypes was 133 ± 35 (n=31) for the wildtype, 121 ± 32 (n=66) for the heterozygous, and 122 ± 36 (n=43) for the homozygous knockout (Figure 4 ). A two-sided Wilcoxon rank sum test indicated no significant difference between any two of these three genotypes (p>0.05). The sizes of all the tumors in 10 mice randomly selected from each genotype were measured to study the possible role of clusterin deficiency in net tumor growth. The average maximum diameters of intestinal tumors are 1.38 ± 0.23 mm for the wildtype, 1.62 ± 0.26 mm for the heterozygote, and 1.52 ± 0.31 mm for the homozygous knockout. Again, a Wilcoxon rank sum test of the size distributions found no significant difference between the wildtype and the homozygous knockout or between the heterozygote and the homozygous knockout (p>0.05). Consequently, the tumor-associated expression of clusterin is not uniquely essential for Min-induced intestinal tumor initiation or growth.
Figure 4.
Clusterin deficiency has no effect on Min tumor multiplicity. The multiplicity of intestinal tumors in Min mice was plotted according to their clusterin genotype. Three groups represented Min mice carrying wildtype clusterin, heterozygous mutants, and homozygous deficiency mutants. A two-sided Wilcoxon sum test indicated no significant difference among these three groups.
Discussion
A number of marker genes for colorectal cancer have been identified from transcript profiling performed with cancer cell lines, human samples, and samples from animal models (Paoni et al. 2003; Leclerc et al. 2004). Understanding the biological significance of these genes in colorectal tumors requires knowledge of their cellular expression patterns from histological analysis. Understanding their potential roles in tumor biology requires genetic manipulation of animal models. Here we have reported the cellular expression patterns of 20 candidate marker genes overexpressed in colorectal tumors nominated by SSH and MA (Table 1). Although they each have significantly elevated levels of expression in Min colonic tumors, their patterns of expression in normal tissue lead to three groups: proliferation zone associated, Paneth cell associated, and tumor associated. All three classes of genes can be detected within the same colonic tumor (data not shown). Their overexpression patterns in human tumors were also confirmed by IHC. Finally, we studied the effect of one candidate gene, clusterin, on Min tumorigenesis. Although clusterin showed strong tumor-associated expression in Min intestinal tumors, ablation of this gene did not measurably affect the multiplicity or growth rate of intestinal tumors.
Using ISH, we successfully analyzed these genes for their RNA expression within murine colorectal tumors. The advantage of this approach is that the gene-specific RNA probes can be conveniently synthesized based on the cDNA information. This approach, however, requires faithful preservation of RNA during tissue collection and processing, which can be problematic in archived human tissues. In contrast, IHC with protein-specific antibodies does not have this limitation. We showed the expression of several candidate genes using IHC in archived human colonic tissues. This approach, however, can be compromised by cross-reaction by the available antibodies. Thus, both IHC and ISH may generate false positives and need rigorous negative controls to ensure specificity. Combining these two approaches on same tissue can greatly reduce the probability of false positives. The most reliable negative control for each approach, however, is to use homozygous-null tissues from mice with a targeted ablation of the gene of interest. We used tumors from clusterin-deficient Min mice to prove that our ISH and IHC assays for clusterin are not compromised by cross-reaction (data not shown).
Mice with a targeted gene ablation provide a central tool to analyze the function of a candidate gene in tumorigenesis. One example is MMP7, a secreted metalloproteinase degrading extracellular matrix (Ii et al. 2006), which displays elevated expression in murine intestinal tumors and in Paneth cells. MMP7-deficient mice carrying the Min mutation develop significantly fewer intestinal tumors than those with wildtype MMP7, indicating that the MMP7 is involved in early tumorigenesis (Wilson et al. 1997). In contrast, clusterin expression is strongly elevated in intestinal tumors from a very early stage (Chen et al. 2003), but we report here that clusterin deficiency alone does not affect either the multiplicity or the growth of Min-induced intestinal tumors. One must consider, however, the dual function of clusterin. The clusterin gene generates two protein products through alternative splicing and glycosylation (Yang et al. 2000). Functionally, the highly glycosylated soluble form (sClu) is anti-apoptotic and pro-tumorigenic, whereas the poorly glycosylated nuclear form (nClu) is pro-apoptotic and anti-tumorigenic (Shannan et al. 2006). Therefore, it is conceivable that ablation of the clusterin gene would remove both products with contrasting functions, so that no overt net effect would be observed.
Because the candidate genes identified in the mouse tumors have similar expression patterns in human tumors, the cellular level expression analysis and functional study in the Min mouse provide useful information for clinical study of colon cancer. Some candidate genes with tumor-associated expression represent secreted proteins, such as clusterin. The signatures of these genes present in the serum of patients may serve as markers for the early detection and diagnosis of this disease. Furthermore, targeted gene deletion will provide a subset of candidate genes that are necessary for tumor initiation, maintenance, and/or progression. The products of these genes could be used as targets of drug intervention for human colorectal cancer. Further studies will emerge from this strategy of coupling the power for discovery of array and cDNA subtraction to the cellular resolution provided by ISH and IHC.
Acknowledgments
This research was supported by Grants R37-CA63677 and U01-CA84227 from the National Cancer Institute to W.F.D. This is publication #3638 from the Laboratory of Genetics, University of Wisconsin–Madison, Madison, WI.
The authors thank our colleagues in the Mouse Models for Human Cancer Consortium for the shared microarray analysis that has provided one starting point for this study. David Threadgill generously provided sections of appropriately fixed AOM tumors for our ISH analysis. Jose Torrealba generously provided sections of human colonic lesions. The Histotechnology Facility of the McArdle Laboratory (Jane Weeks, leader) provided sections of high quality for this study. Cheri Pasch, Ryan Burch, and Kathy Krentz provided fastidious assistance in ISH and Dawn Albrecht in the maintenance of pedigreed mouse kindreds. Finally, we thank members of the Dove laboratory for helpful discussion and especially Linda Clipson for critical input and skilled execution in the generation of this manuscript.
References
- American Cancer Society (2007) Cancer Facts and Figures 2007. Atlanta, American Cancer Society
- Bedell MA, Largaespada DA, Jenkins NA, Copeland NG (1997) Mouse models of human disease. Part II: recent progress and future directions. Genes Dev 11:11–43 [DOI] [PubMed] [Google Scholar]
- Buckhaults P, Rago C, St Croix B, Romans KE, Saha S, Zhang L, Vogelstein B, et al. (2001) Secreted and cell surface genes expressed in benign and malignant colorectal tumors. Cancer Res 61:6996–7001 [PubMed] [Google Scholar]
- Chen J, Chang S, Duncan SA, Okano HJ, Fishell G, Aderem A (1996) Disruption of the MacMARCKS gene prevents cranial neural tube closure and results in anencephaly. Proc Natl Acad Sci USA 93:6275–6279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Halberg RB, Ehrhardt WM, Torrealba J, Dove WF (2003) Clusterin as a biomarker in murine and human intestinal neoplasia. Proc Natl Acad Sci USA 100:9530–9535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich WF, Lander ES, Smith JS, Moser AR, Gould KA, Luongo C, Borenstein N, et al. (1993) Genetic identification of Mom-1, a major modifier locus affecting Min-induced intestinal neoplasia in the mouse. Cell 75:631–639 [DOI] [PubMed] [Google Scholar]
- Dove WF, Cormier RT, Gould KA, Halberg RB, Merritt AJ, Newton MA, Shoemaker AR (1998) The intestinal epithelium and its neoplasms: genetic, cellular, and tissue interactions. Philos Trans R Soc Lond B Biol Sci 353:915–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink D, Fazli L, Aronow B, Gleave ME, Ong CJ (2006) Clusterin is not essential for androgen-regulated involution and regeneration of the normal mouse prostate. Prostate 66:1445–1454 [DOI] [PubMed] [Google Scholar]
- Hugh TJ, Dillon SA, O'Dowd G, Getty B, Pignatelli M, Poston GJ, Kinsella AR (1999) β-catenin expression in primary and metastatic colorectal carcinoma. Int J Cancer 82:504–511 [DOI] [PubMed] [Google Scholar]
- Ii M, Yamamoto H, Adachi Y, Maruyama Y, Shinomura Y (2006) Role of matrix metalloproteinase-7 (matrilysin) in human cancer invasion, apoptosis, growth, and angiogenesis. Exp Biol Med (Maywood) 231:20–27 [DOI] [PubMed] [Google Scholar]
- Kaiser S, Park YK, Franklin JL, Halberg RB, Yu M, Jessen WJ, Freudenberg J, et al. (2007) Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome Biol 8:R131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc D, Deng L, Trasler J, Rozen R (2004) ApcMin/+ mouse model of colon cancer: gene expression profiling in tumors. J Cell Biochem 93:1242–1254 [DOI] [PubMed] [Google Scholar]
- Li J, Aderem A (1992) MacMARCKS, a novel member of the MARCKS family of protein kinase C substrates. Cell 70:791–801 [DOI] [PubMed] [Google Scholar]
- Lobach DF, Rochelle JM, Watson ML, Seldin MF, Blackshear PJ (1993) Nucleotide sequence, expression, and chromosomal mapping of Mrp and mapping of five related sequences. Genomics 17:194–204 [DOI] [PubMed] [Google Scholar]
- Moser AR, Pitot HC, Dove WF (1990) A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science 247:322–324 [DOI] [PubMed] [Google Scholar]
- Muro S, Takemasa I, Oba S, Matoba R, Ueno N, Maruyama C, Yamashita R, et al. (2003) Identification of expressed genes linked to malignancy of human colorectal carcinoma by parametric clustering of quantitative expression data. Genome Biol 4:R21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoni NF, Feldman MW, Gutierrez LS, Ploplis VA, Castellino FJ (2003) Transcriptional profiling of the transition from normal intestinal epithelia to adenomas and carcinomas in the APCMin/+ mouse. Physiol Genomics 15:228–235 [DOI] [PubMed] [Google Scholar]
- Powell SM, Zilz N, Beazer-Barclay Y, Bryan TM, Hamilton SR, Thibodeau SN, Vogelstein B, et al. (1992) APC mutations occur early during colorectal tumorigenesis. Nature 359:235–237 [DOI] [PubMed] [Google Scholar]
- Shannan B, Seifert M, Leskov K, Willis J, Boothman D, Tilgen W, Reichrath J (2006) Challenge and promise: roles for clusterin in pathogenesis, progression and therapy of cancer. Cell Death Differ 13:12–19 [DOI] [PubMed] [Google Scholar]
- Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, et al. (2006) The consensus coding sequences of human breast and colorectal cancers. Science 314:268–274 [DOI] [PubMed] [Google Scholar]
- Su LK, Kinzler KW, Vogelstein B, Preisinger AC, Moser AR, Luongo C, Gould KA, et al. (1992) Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science 256:668–670 [DOI] [PubMed] [Google Scholar]
- van Es JH, Jay P, Gregorieff A, van Gijn ME, Jonkheer S, Hatzis P, Thiele A, et al. (2005) Wnt signalling induces maturation of Paneth cells in intestinal crypts. Nat Cell Biol 7:381–386 [DOI] [PubMed] [Google Scholar]
- Wegele H, Muller L, Buchner J (2004) Hsp70 and Hsp90–a relay team for protein folding. Rev Physiol Biochem Pharmacol 151:1–44 [DOI] [PubMed] [Google Scholar]
- Williams NS, Gaynor RB, Scoggin S, Verma U, Gokaslan T, Simmang C, Fleming J, et al. (2003) Identification and validation of genes involved in the pathogenesis of colorectal cancer using cDNA microarrays and RNA interference. Clin Cancer Res 9:931–946 [PubMed] [Google Scholar]
- Wilson CL, Heppner KJ, Labosky PA, Hogan BL, Matrisian LM (1997) Intestinal tumorigenesis is suppressed in mice lacking the metalloproteinase matrilysin. Proc Natl Acad Sci USA 94:1402–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CL, Ouellette AJ, Satchell DP, Ayabe T, Lopez-Boado YS, Stratman JL, Hultgren SJ, et al. (1999) Regulation of intestinal alpha-defensin activation by the metalloproteinase matrilysin in innate host defense. Science 286:113–117 [DOI] [PubMed] [Google Scholar]
- Yang CR, Leskov K, Hosley-Eberlein K, Criswell T, Pink JJ, Kinsella TJ, Boothman DA (2000) Nuclear clusterin/XIP8, an x-ray-induced Ku70-binding protein that signals cell death. Proc Natl Acad Sci USA 97:5907–5912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhou W, Velculescu VE, Kern SE, Hruban RH, Hamilton SR, Vogelstein B, et al. (1997) Gene expression profiles in normal and cancer cells. Science 276:1268–1272 [DOI] [PubMed] [Google Scholar]