Abstract
The induction of porcine cytokines, which are believed to be important for the regulation of T helper (Th)1- and Th2-specific immune responses of pigs, was analysed after in vitro restimulation with a herpesvirus, Suid herpes 1 (pseudorabies virus [PRV]), in peripheral blood mononuclear cells (PBMC). To this end, quantitative, competitive reverse transcription–polymerase chain reaction (RT–qcPCR) was established using constructed heterologous DNA MIMICS, which contain cytokine- or glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-specific primer-binding sites. This is a simple method that allows reliable determination of the differing regulation of cytokine mRNAs specific for porcine interleukin (IL)-2, -4 and -10, interferon gamma (IFN-γ) and the housekeeping gene, GAPDH, as an endogenous control. PBMC derived from naive (innate response) and PRV-primed (memory response) outbred swine were analysed comparatively. The results demonstrated that restimulation with PRV significantly enhanced the transcription of Th1-type cytokines (IL-2 and IFN-γ) but not of Th2-type cytokines (IL-4 and IL-10). This virus-specific cytokine response was only found with PBMC from swine protected against lethal PRV challenge infection, but not with naive PBMC or with PBMC from pigs immunized with plasmid DNA encoding PRV glycoprotein gC. Notably, PBMC derived from immune and naive pigs constitutively produced relatively high amounts of IL-10-specific mRNA, exceeding that of GAPDH mRNA, independently of the addition of viral antigen or the mitogen concanavalin A (Con A). The results of this work should help to provide a better understanding of the effector cell/cytokine network response to infection with, or vaccination against, PRV. Additionally, the simple, reliable and sensitive RT–qcPCR, when used to determine the porcine cytokine pattern, might be of prognostic value for the induction of protective immunity.
Introduction
Pseudorabies virus (PRV), a member of the Alphaherpesvirinae, is the causative agent of Aujeszky's disease, an economically important disease of swine. Vaccination of pigs is practised in many countries, using live and inactivated vaccines. Although vaccines can prevent symptoms of the disease, they do not eliminate the virus from the infected host and do not prevent the establishment of a latent infection (reviewed in ref. 1). More recently, the use of plasmid DNAs (encoding glycoproteins of PRV) for DNA vaccination of pigs has been reported with varying success, particularly regarding protection of the animals against lethal challenge with highly virulent PRV strains.2–5
One important prerequisite for the improvement of anti-herpesviral vaccines in general, and against PRV in particular, is a better knowledge of immunological parameters relevant for protection. For instance, the presence and activation of long-lived memory B- and T-cell responses is important to confer long-lasting immunity against herpesviruses.6 Specific virus-neutralizing serum antibodies are believed to play a substantial role in controlling PRV infection, and recently it has been demonstrated that a humoral immune response directed against glycoprotein gC can interfere directly with the attachment of PRV to cells.7 However, it has also been shown that high titres of circulating antibodies are not solely responsible for protective immunity.1,8 Cell-mediated immunity is regarded as a major protective effector mechanism against virus infections,6,9 including herpes simplex virus (HSV) or PRV.10–13 Pigs infected with or vaccinated against PRV produce specific cytotoxic T cells, which are major histocompatibility complex (MHC) unrestricted as well as MHC class I restricted.12,14 CD4+ T helper (Th) cells have been found among porcine T lymphocytes with anti-PRV activity.15 The CD4+ CD8+ extrathymic T cells, which are unique to the porcine immune system, contain cells responsive to PRV as recall antigen that are characteristic of memory T cells.16–18 This virus-specific secondary response was found to be MHC class II restricted, requiring the CD4 molecule for recognition, and the glycoprotein gC, a major immunogen of PRV, was found to be involved in the priming of PRV-specific memory Th cells.7,17
It is well documented that cytokines and chemokines, secreted by macrophages and T lymphocytes, play a crucial role in the initiation and maintenance of antiviral immune responses (reviewed in ref. 19). Two different Th cell subsets (Th1 and Th2), which differ from each other in their cytokine profile, are described for mouse, human and bovine T lymphocytes.20–22 Th1-type cytokines, such as interleukin (IL)-2 and interferon-γ (IFN-γ), stimulate cytotoxic T lymphocytes (CTLs) and B cells, whereas IL-4 and IL-10 predominately promote B-cell activity.23 The Th1-type of immune response is believed to limit viral replication and to facilitate resolution of viral infections.9,24 It was demonstrated in mice that a Th1-type cytokine-mediated immune response was crucial for the prevention of lethal HSV infection, whereas a Th2-type immune response aggravated the disease.25,26 Using mouse models for PRV, the induction of a Th1-type of antiviral immune response and its importance for protective immunity have been reported.27,28
Information on the porcine cytokine profile in PRV-infected or vaccinated swine, the natural host of PRV, is very scarce owing to the limited availability of appropriate immunological and biological reagents. To date, indirect assays to measure cytokine activity indicate secretion of IL-2 and IFN-γ after in vitro restimulation of PRV-primed porcine lymphocytes.8,17,29,30 An excellent alternative for the quantitative assessment of cytokine gene expression is competitive reverse transcription–polymerase chain reaction (RT–PCR),31,32 also described for swine.33–36 In the present report we determined cytokine gene expression in peripheral blood mononuclear cells (PBMC) of naive (innate response) and PRV-primed (memory response) outbred swine after in vitro stimulation with PRV or concanavalin A (Con A). To achieve this, a reliable and simple RT–qc (quantitative, competitive) PCR was developed for accurate quantification of porcine IL-2, -4 and -10, and IFN-γ. Animals vaccinated with a live vaccine were fully protected against challenge infection with a lethal dose of PRV, and the derived PBMC exhibited significantly increased IL-2 and IFN-γ expression upon in vitro restimulation with PRV. This induction was PRV specific and did not occur with PBMC from naive animals. In contrast, transcription of neither IL-2 nor IFN-γ was stimulated by PRV after exposure of PBMC derived from piglets that were vaccinated with a plasmid encoding gC of PRV and not protected against lethal virus challenge. Induction of IL-4 and IL-10 was absent after restimulation of PBMC with PRV, whereas treatment with the polyclonal T-cell activator Con A increased transcription of all cytokines tested, except IL-10. Notably, in all PBMC samples analysed, a relatively high basal transcription of IL-10 was observed, which even exceeded that of the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Improved knowledge of the cytokine profiles will help to elucidate the contribution of different porcine T-lymphocyte subsets in protective immunity and therefore assist in improving the development and delivery of protective vaccines, as exemplified here for PRV infection in pigs. Moreover, better characterization of the cytokine response against primary and secondary viral infection might also be of prognostic value for the protective quality of vaccines in general.
Similarly to herpesvirus infection in humans and mice, the results presented in this work indicate that in successful protection of swine against PRV infection a Th1-type immune response prevails, which was not found after gC DNA immunization.
Materials and methods
Virus propagation and chemical inactivation of PRV
The porcine kidney cell line, PSEK, was used for propagation of the PRV strain, Phylaxia, as described previously.7 For inactivation, the virus lysate (108 plaque-forming units [PFU] per ml) was incubated with 0·08% (v/v) β-propiolactone (Sigma-Aldrich Chemie GmbH, Munich, Germany) on ice for 30 min with occasional shaking. Thereafter, the mixture was shaken at 37° for 4 hr (taking care to maintain the pH between 7·0 and 7·4) and then incubated at 4° for an additional 48 hr without shaking. The resultant precipitate was removed by centrifugation at 400 g for 15 min, and the supernatant containing PRV was assayed for successful inactivation of virus by inoculation of PSEK cells.
Animals and immunization
Pigs (German landrace; weight ≈ 25 kg) were immunized intramuscularly (i.m.) with 106 of a 50% tissue culture infective dose (TCID50) of PRV live vaccine (Nobi-Porvac live; Intervet, Boxmeer, the Netherlands) and boosted 4 weeks later (105 TCID50). Two months later, all animals survived an intranasal challenge infection with 1–2 × 105 PFU of the highly virulent PRV strain, NIA-3. All immune sera displayed PRV-specific antibody titres between 1 : 2000 and > 1 : 10 000, as determined by enzyme-linked immunosorbent assay (ELISA) and complement-independent virus-neutralizing antibody titres of 1 : 64–1 : 480 (data not shown).
Animals 9, 10 and 15 were injected intradermally (i.d.) in the pinna of both ears with 50 µg of plasmid DNA containing the gC gene (gC-CMV) (25 µg of DNA in 0·1 ml of phosphate-buffered saline [PBS] was administered at each site using a 22-gauge needle). Two booster immunizations were performed at subsequent 4-week intervals by i.d. inoculation of a further 50 µg of gC-CMV, and 4 weeks after the third immunization the animals were intranasally challenge infected (with 1–2 × 105 PFU of NIA-3). Plasmid gC-CMV was kindly provided by T. C. Mettenleiter (Federal Research Centre For Virus Diseases of Animals, Insel Riems, Germany) and has been described previously.3
Isolation and in vitro stimulation of PBMC
Heparinized blood was collected from the jugular vein, and PBMC were prepared by Ficoll gradient centrifugation and cultured as described previously.7 In vitro stimulation was performed with 5 µg/ml of Con A (Sigma-Aldrich Chemie GmbH) with PRV (strain Phylaxia) or with bovine herpesvirus 1 (BHV-1) (vaccine strain Difivac; Bayer AG, Leverkusen, Germany) at a multiplicity of infection (MOI) of 1·0.
RNA extraction
Total RNA was isolated from PBMC with TRIzol reagent (Gibco-BRL Life Technologies, Karlsruhe, Germany), according to the manufacturer's instructions, and resuspended in 20 µl of sterile bidistilled water containing 0·1% (v/v) diethyl-pyrocarbonate (DEPC; Sigma-Aldrich Chemie GmbH). RNA was quantified spectrophotometrically and tested for integrity by using denaturing agarose-gel electrophoresis.
RT of total RNA and PCR
Total cellular RNA from PBMC was heated at 70° for 10 min, chilled on ice and then incubated for 10 min at room temperature in a total volume of 20 µl containing 2 µg of RNA, 0·25 µg of random hexamer primers (Invitrogen BV, Groningen, the Netherlands), 0·5 mm deoxynucleotide triphosphates (dNTPs; Amersham Pharmacia Biotech, Freiburg, Germany), 1× first-strand buffer (Gibco-BRL Life Technologies), 10 mm dithiothreitol (Gibco-BRL Life Technologies) and 20 U of ribonuclease inhibitor (RNaseOUT; Gibco-BRL Life Technologies). After addition of 200 U of reverse transcriptase (Superscript II, Gibco-BRL Life Technologies), cDNA was synthesized at 42° for 1 hr followed by heat inactivation at 70° for 10 min.
Oligonucleotides used as primers for PCR amplification of cDNA specific for porcine GAPDH, IL-2, IFN-γ, IL-4 and IL-10 are listed in Table 1. PCR was performed, for 35 cycles, in a total volume of 50 µl containing 20 mm Tris-HCl (pH 8·4), 50 mm KCl, 1·5 mm MgCl2, 30 ng of each primer, 0·02 mm dNTPs (Amersham Pharmacia Biotech), 1 µl of the cDNA reaction and 0·5 µl of Taq polymerase (5 U/µl; Gibco-BRL Life Technologies) in a TRIO-Thermoblock (Biometra, Göttingen, Germany). Each PCR cycle comprised 80 seconds of denaturation at 96°, 90 seconds of annealing at 56° (64° for the IL-4-specific PCR), a 90-second extension at 72° and a final extension at 72° for 10 min. The reaction products (15 µl) were separated in 2% agarose gels containing 0·3 µg/ml ethidium bromide. Specific PCR products were cloned into the pCR™II cloning vector (Invitrogen BV), according to the manufacturer's instructions, and then sequenced using an automatic DNA sequencing apparatus (ABI model 377 sequencing system; Applied Biosystems Division PE/ABI, Foster City, CA) and the ABI PRISM™ Dye-Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems GmbH, Perkin-Elmer Corporation, Weiterstadt, Germany).
Table 1.
Primers used for quantitative, competitive reverse transcription–polymerase chain reaction (RT–qcPCR)
| Product size (bp) | ||||
|---|---|---|---|---|
| Target | Primer sequence (5′−3′) | Target | MIMIC | GenBank acc. no. |
| IL-2 | Forward GCACCTACTTCAAGCTCTAC | 387 | 431 | X56750 |
| Reverse GATGCTTTGACAAAAGGTAATC | ||||
| IL-4 | Forward TATTCATGGGTCTCACCTCCCA | 337 | 432 | X68330 |
| Reverse TTGGCTTCATGCACAGAACAG | ||||
| IL-10 | Forward TACCTGGGTTGCCAAGCCTT | 523 | 431 | L20001 |
| Reverse TTCACAGAGAGGCTCGGTAAAT | ||||
| IFN-γ | Forward ATTTTGAAGAATTGGAAAGAGG | 368 | 433 | X53085 |
| Reverse AAATTCAAATATTGCAGGCAGG | ||||
| GAPDH | Forward CCTTCATTGACCTCAACTACAT | 400 | 432 | U48832 |
| Reverse CCAAAGTTGTCATGGATGACC | ||||
GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IFN-γ, interferon-γ; IL, interleukin.
Quantitative, competitive PCR
A heterologous competitor DNA (MIMIC) was generated from the Escherichia coli lacZ gene by PCR using plasmid pCH110 (Amersham Pharmacia Biotech) as a template. Composite primers, consisting of the target gene-specific primer sequence (Table 1) linked to a lacZ gene-specific sequence, were synthesized. Therefore, all forward primers contained at their 3′ end the additional sequence 5′-CTCGTTGCTGCATAAACC-3′, and all reverse primers 5′-GCAGACCATTTTCAATCC-3′. The PCR was performed as described above, but using an annealing temperature of 50°. The resulting PCR products were diluted 250-fold and used for a second round of amplification in the presence of the target-specific primers. The resulting MIMICs were purified using the High Pure PCR-Product Purification Kit (Roche Molecular Biochemicals, Mannheim, Germany), and the DNA concentration was determined. The molar quantity of each MIMIC resulted from the calculation that 1 ng of DNA of a size of 300 bp equals 5 × 103 attomoles (amol).
Serial, 10-fold dilutions of each MIMIC were placed into different test tubes and amplified with constant amounts (1 µl) of cDNA. The resulting PCR products were separated in an ethidium bromide-stained 2% agarose gel and photographed using the DC120 Zoom Digital Camera (Eastman Kodak Company, Rochester, NY). Computer-directed densitometry was performed using 1D Image Analysis Software (Eastman Kodak Company) to determine the maximum fluorescence intensity of each DNA band. The molar ratio between target and MIMIC-specific PCR products was calculated for each reaction using the equation:
The log10 value of the molar ratio was plotted against the log10 value of amol of MIMIC present in each reaction. This regression plot yielded a linear graph, and from the intercept on the x-axis, the point of molar equivalence was determined, representing the relative amount of target cDNA. Quantification of the cDNA specific for the housekeeping gene, GAPDH, was used as a control for any sample-to-sample variation in quantity of RNA and variation of RT and PCR. Therefore, the values obtained for individual stimulated and non-stimulated PBMC samples were corrected by their GAPDH ratios. This finally allowed the quantitative comparison of changes in the cytokine profiles and was expressed as the induction coefficient of cytokine transcription (ICT).
Results
Establishment of RT–PCR for specific detection of porcine cytokine transcripts
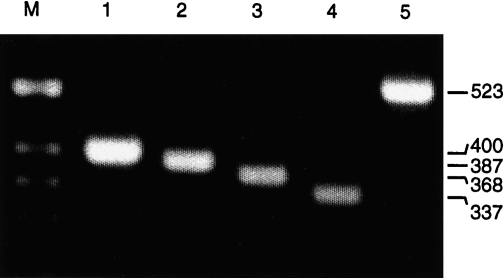
Stimulation of porcine PBMC with the polyclonal T-cell activator Con A allows the detection of various cytokines, including IL-2, IFN-γ, IL-4 and IL-10.34 Therefore, RT–PCR was established and optimized initially with PBMC derived from a naive pig (pig 1), which were stimulated in vitro for 24 hr with Con A. The RT–PCR resulted in specific amplification products of the expected size: 400 bp for GAPDH, 387 bp for IL-2, 368 bp for IFN-γ, 337 bp for IL-4 and 523 bp for IL-10 (Fig. 1). To prevent amplification of cytokine-specific genomic DNA, the chosen primers were specific for two different exons of each cytokine gene. Although the GAPDH-specific primers are positioned in the same exon, GAPDH-specific genomic DNA was not amplified. Reverse transcription in the absence of reverse transcriptase or when using RNase A-digested RNA did not result in a GAPDH-specific PCR product, whereas DNase I treatment of the RNA did not affect the RT–PCR (not shown). In addition, the identity of all resulting RT–PCR products was further confirmed by sequence analysis of the cloned PCR fragments (results not shown).
Figure 1.
Reverse transcription–polymerase chain reaction (RT–PCR) specific for porcine cytokines and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Total RNA was isolated from concanavalin A (Con A)-stimulated (24 hr) peripheral blood mononuclear cells (PBMC) of a naive pig and used for RT–PCR specific for: GAPDH (lane 1), interleukin (IL)-2 (lane 2), interferon-γ (IFN-γ) (lane 3), IL-4 (lane 4) and IL-10 (lane 5). The PCR products were separated in a 2% agarose gel; the sizes (in bp) for each specific product are indicated. M, molecular-weight markers.
The sensitivity of the cytokine- and GAPDH-specific PCR amplification was determined by using serial dilution of known amounts of each cloned PCR product as template. The assays revealed that 10−6 pm of IL-4-, 10−5 pm of GAPDH-, IL-2- and IL-10-, and 10−3 pm of IFN-γ-specific cDNA could be detected in ethidium bromide-stained gels (results not shown).
Cytokine profile in Con A-activated PBMC
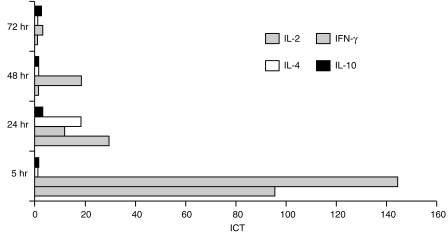
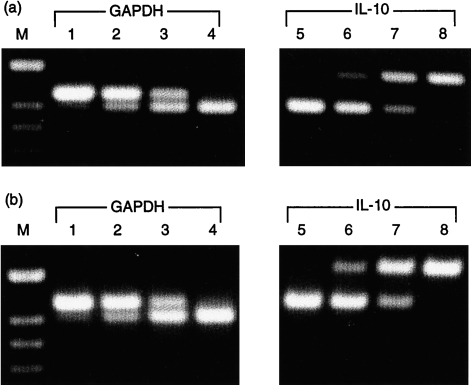
To assess the stimulation and amplification of cytokine mRNA, PBMC from non-infected pig 1 were cultured in the presence or absence of Con A, and total RNA was isolated from 1 × 107 cells after 5, 24, 48 and 72 hr. The transcription rate of GAPDH, IL-2, IFN-γ, IL-4 and IL-10 was analysed by RT–qcPCR, and the ICT was determined as described in the Materials and methods. The kinetic profile of cytokine induction in Con A-activated porcine PBMC is illustrated in Fig. 2. Maximal induction of the IL-2- and IFN-γ-specific transcription (95·8-fold and 144·0-fold, respectively) was observed as early as 5 hr after addition of Con A to the PBMC cultures. Thereafter, the ICT of IL-2 and IFN-γ gradually decreased to basal levels (as produced by non-stimulated cells) after 48 hr (IL-2) and 72 hr (IFN-γ) of culture. Elevated transcription of IL-4 was found only after 24 hr of stimulation with Con A, showing an ICT of at least 18·0, and was essentially undetectable at earlier or later time-points and not detected in non-stimulated cells (Fig. 2). Con A caused no significant increase of IL-10 transcription during the time-period tested (Fig. 2). Notably, gel analysis of the RT–qcPCR products indicated the presence of relatively high amounts of IL-10 mRNA in non-stimulated PBMC. Molar equivalence was reproducibly found in the presence of ≈ 1–10 amol IL-10 MIMIC using non-stimulated (Fig. 3a) or Con A-stimulated PBMC (Fig. 3b). For GAPDH, equivalence was reached with ≈ 10- to 20-fold lower concentrations of MIMIC (Fig. 3, lanes 1–4). This was not the result of a higher sensitivity of the IL-10-specific PCR compared to the other PCRs (as described above), and was found with all test PBMC samples derived from the different pigs (also discussed below).
Figure 2.
Kinetics of cytokine induction in porcine peripheral blood mononuclear cells (PBMC) during treatment with concanavalin A (Con A). PBMC derived from a naive pig were stimulated in vitro with Con A, and cytokine transcription was quantified by quantitative, competitive reverse transcription–polymerase chain reaction (RT–qcPCR) at the indicated time-points. By comparison with the non-stimulated PBMC, the induction coefficients of transcription (ICT) were determined as described in the Materials and methods. The highest ICT were found for interleukin (IL)-2 and interferon-γ (IFN-γ) after 5 hr of stimulation with Con A, whereas IL-4 transcription was induced only after 24 hr. IL-10 transcription did not show a significant increase at any of the time-points tested.
Figure 3.
Quantification of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and interleukin (IL)-10 mRNA. Quantitative, competitive reverse transcription–polymerase chain reaction (RT–qcPCR) was performed with (a) non-stimulated and (b) concanavalin A (Con A)-stimulated (24 hr) peripheral blood mononuclear cells (PBMC) from a naive pig. For PCR, cDNA and a 10-fold dilution of MIMIC were added together in concentrations ranging from 101 to 10−2 amol for GAPDH (lanes 1–4, upper bands), and from 102 to 10−1 amol for IL-10 (lanes 5–8, lower bands). In both Con A- and non-stimulated PBMC, molar equivalence was obtained in the presence of ≈ 100−101 amol IL-10-specific MIMIC (lane 7, both panels) and 10−1 amol GAPDH-specific MIMIC (lane 3, both panels). M, molecular-weight markers.
To examine the variability of cytokine induction among individual outbred pigs, PBMC derived from nine additional pigs were stimulated with Con A for 24 hr and analysed. As summarized in Table 2, the ICT for IL-2 ranged from 10·4 to 66·9 (average 24·4), for IFN-γ from 3·6 to 45·4 (average 20·0) and for IL-4 from 12·0 to > 54·0 (average 22·0). A more exact determination of the ICT of IL-4 was not possible because IL-4 mRNA could be not detected in any of the non-stimulated PBMC. In no case was a significant induction of the basal IL-10 transcription, which never increased more than 2·8-fold (average 1·6), found after Con A stimulation (Table 2).
Table 2.
Induction coefficients of cytokine transcription (ICT) in porcine peripheral blood mononuclear cells (PBMC)
| Con A | PRV (inactivated) | |||||||
|---|---|---|---|---|---|---|---|---|
| Animals | IL-2 | IFN-γ | IL-4 | IL-10* | IL-2 | IFN-γ | IL-4 | IL-10 |
| Pig 1 (naive) | 29·6 | 11·9 | > 18·0 | + | NT† | NT | NT | NT |
| Pig 2 (naive) | 10·4 | 23·4 | > 12·0 | + | 0·6 | 0·9 | –‡ | + |
| Pig 3 (naive) | 15·3 | 19·9 | > 27·0 | + | 3·4 | 1·4 | – | + |
| Pig 4 (naive) | 12·2 | 6·4 | > 54·0 | + | 3·4 | 1·6 | – | + |
| Pig 5 (immune) | 11·9 | 13·2 | > 10·0 | + | 59·6 | 40·0 | – | + |
| Pig 6 (immune) | 17·1 | 45·0 | > 33·0 | + | 124·4 | 63·0 | – | + |
| Pig 7 (immune) | 13·1 | 45·4 | ++§ | + | 42·1 | 34·3 | – | + |
| Pig 9 (gC-DNA) | 30·7 | 6·6 | ++ | + | 2·1 | 2·6 | – | + |
| Pig 10 (gC-DNA) | 66·9 | 3·6 | ++ | + | 3·5 | 1·6 | – | + |
| Pig 15 (gC-DNA) | 12·7 | > 4·8 | ++ | + | 0·5 | – | – | + |
+ indicates the high constitutive transcription of interleukin (IL)-10, which did not increase significantly (also see the text).
Not tested.
Not detectable.
The clear induction of IL-4 is indicated by ++, but the exact ICT was not determined.
gC, PRV glycoprotein gC; IFN-γ, interferon-γ; PRV, pseudorabies virus.
Estimation of molar amounts of cytokine mRNA
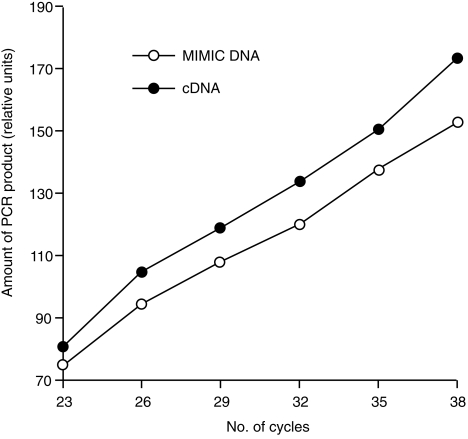
The results described above indicate varying basal levels of the different cytokine mRNAs in non-stimulated PBMC: particularly high for IL-10 and very low for IL-4. The RT–qcPCR used in this work allows accurate determination of relative differences in cytokine gene expression, but not necessarily the correct quantification of molar concentrations of the synthesized target cDNA reflecting the amount of mRNA molecules present in each sample. For such an estimation, target and MIMIC must share equal amplification kinetics. This was tested with identical amounts (10 amol) of the cloned target cDNA and the respective MIMIC as templates in the same PCR. Five microlitres of the PCR reaction was collected after 23, 26, 29, 32, 35 and 38 cycles and separated by gel electrophoresis. The amount of PCR products specific for MIMIC and target were quantified by densitometry and plotted against the number of cycles. Representative results are shown for the simultaneous amplification of GAPDH-specific cDNA and the GAPDH MIMIC (Fig. 4). The almost parallel slope of both curves indicates that both templates were amplified to approximately equal molar amounts. This could also be demonstrated for the other target cDNAs and their corresponding MIMIC, which were all amplified with nearly identical kinetics (data not shown). Therefore, the actual number of mRNA molecules present in each sample could be estimated from the RT–qcPCR results. Because efficiency of cDNA synthesis is probably less than 100%, the estimated values represent the minimum number of mRNA molecules present in each sample. Using RNA isolated from non-stimulated PBMC derived from the different naive pigs (pigs 1–4) ≈ 105.7 copies of IL-10 mRNA were found per µg of total cellular RNA, but 25-fold less GAPDH mRNA (104.3 copies/µg of total RNA). Compared with IL-2 and IFN-γ (103.3 and 103.2 copies of mRNA/µg of total RNA, respectively) the basal transcription rate of IL-10 in naive PBMC was, on average, 250-fold and 300-fold higher, respectively. In summary, these results verified the presumption that IL-10 mRNA was constitutively transcribed in relatively high amounts in porcine PBMC during 24 hr of culture.
Figure 4.
Kinetics of amplification of glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-specific target cDNA and MIMIC. Identical amounts of both templates (10 amol) were co-amplified, parts of the reaction separated in a 2% agarose gel at the indicated number of polymerase chain reaction (PCR) cycles, and the amount of each PCR product determined densitometrically. The slope of both curves indicated very similar efficiency of amplification for the target cDNA (filled circles) and the MIMIC (open circles).
Th1-type cytokine response in PRV-primed PBMC
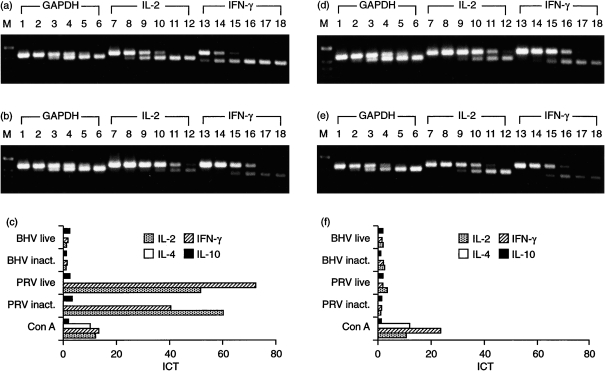
PBMC from naive and PRV-vaccinated, challenge-infected pigs were stimulated in vitro for 24 hr with inactivated PRV (at a MOI of 1·0 before virus inactivation) and the induced cytokine profiles were compared. As controls, cells were left untreated or were stimulated for 24 hr with Con A. Figure 5 represents results obtained with PBMC from an immune animal (pig 5; Fig. 5a, 5b, 5c) and from a naive animal (pig 2, Fig. 5d, 5e, 5f). Gel analysis of the RT–qcPCR products demonstrated that for GAPDH, molar equivalence was obtained in the presence of ≈ 10−1 amol MIMIC (Fig. 5a, 5b, 5d, 5e, lane 4), likewise using PRV-stimulated (Fig. 5a, 5d) or non-stimulated (Fig. 5b, 5e) PBMC. This indicated comparable RNA quality and efficiency of RT–qcPCR for the different PBMC preparations. After restimulation of PBMC of the primed animal with inactivated PRV, the addition of 10−1 amol IL-2 MIMIC and 100 amol IFN-γ MIMIC, respectively, led to molar equivalence (Fig. 5a, lanes 9 and 14). In contrast, RT–qcPCR with RNA from non-stimulated PBMC of the same animal showed equivalence of between 10−3 and 10−4 amol IL-2 MIMIC (Fig. 5b, lanes 11 and 12) and of 10−2 amol IFN-γ MIMIC (Fig. 5b, lane 16). Densitometric analysis of the gel-separated RT–qcPCR products and subsequent calculation of the ICT revealed that restimulation with inactivated PRV increased IL-2 transcription nearly 60-fold and IFN-γ transcription 40-fold (Fig. 5c). Very similar results were obtained with PBMC isolated from two other PRV-immune pigs (pigs 6 and 7). Again, in vitro restimulation with inactivated PRV induced the transcription of IL-2 and IFN-γ to a higher extent than Con A (Table 2). In contrast, PBMC derived from naive animals (pigs 2–4) and stimulated with PRV did not show significantly elevated transcription of IL-2 or IFN-γ (Fig. 5d, 5e, 5f; Table 2).
Figure 5.
Comparative cytokine profiles of peripheral blood mononuclear cells (PBMC) from immune and naive pigs. PBMC were stimulated in vitro with inactivated (inact.) or live pseudorabies virus (PRV), bovine herpesvirus 1 (BHV-1), concanavalin A (Con A) or left unstimulated for 24 hr. Results of quantitative, competitive reverse transcription–polymerase chain reaction (RT–qcPCR) are shown for one immune animal (pig 5) (a), (b) and (c) and one naive animal (pig 2) (d) and (e). The RT–qcPCR products obtained from PRV-stimulated (a and d) and non-stimulated (b and e) PBMC were separated in a 2% agarose gel; PCR was performed in the presence of 102−10−3 amol glyceraldehyde-3-phosphate dehydrogenase (GAPDH) MIMIC (lanes 1–6, upper DNA bands), 101−10−4 amol IL-2 MIMIC (lanes 7–12, upper DNA bands) or interferon-γ (IFN-γ) MIMIC (lanes 13–18, upper DNA bands), respectively. Calculation of the induction coefficients of transcription (ICT), as described in the Materials and methods, is shown in (c) and (f), which demonstrates that T helper 1 (Th1)-type cytokines are specifically induced by PRV only after restimulation of PBMC from the immune pig (a), (b) and (c), which surpassed the ICT found after 24 hr of stimulation with Con A. M, molecular-weight markers.
Upon stimulation with PRV, IL-4-specific RNA was not detected in PBMC from either naive or immune animals (Fig. 5c, 5f), although this PCR was the most sensitive of all the PCR tests. Stimulation of the same PBMC with Con A, however, resulted in induction of IL-4 (Fig. 5c, 5f; Table 2). Similarly to stimulation with Con A, IL-10 transcription was not stimulated by PRV, either in PBMC derived from naive pigs or from PRV-immune pigs (Fig. 5c, 5f; Table 2). In all cases, molar equivalence with target cDNA occurred by the addition of ≈ 10 amol of IL-10 MIMIC after RT–qcPCR.
The results obtained using inactivated PRV for restimulation indicated that viral infection and replication was not necessary for stimulation of IL-2 and IFN-γ. This result could be confirmed using live PRV (MOI 1·0) as recall antigen. Very similar ICT were found for IL-2 (51·3) and IFN-γ (71·8) as compared to restimulation with inactivated PRV of PBMC from the PRV-immune animal (pig 5). Furthermore, live PRV did not stimulate the transcription of IL-4 and IL-10 (Fig. 5c, 5f). Finally, the PRV-specific induction of IL-2 and IFN-γ in immune PBMC was confirmed by exposure of the cells to BHV-1 (at a MOI of 1·0), a closely related alphaherpesvirus. Neither live nor inactivated BHV-1 resulted in a significant increase of ICT for any cytokine tested (Fig. 5c, 5f).
Cytokine response after gC DNA immunization
To examine the cytokine response after DNA immunization, PBMC from three pigs (animals 9, 10 and 15) were analysed after three vaccinations with the gC-expressing DNA plasmid. As summarized in Table 2, transcription of neither IL-2 and IFN-γ nor of IL-4 and IL-10 was induced after in vitro restimulation with PRV. As a control, treatment of PBMC with Con A led to an approximately four- to sevenfold increase in transcription of IFN-γ and a 13- to 67-fold enhanced transcription of IL-2, comparable to the results found with Con A-stimulated PBMC from the other pigs (Table 2). At the time of investigation, the animals had developed a weak humoral immune response against PRV, as reported previously.2,3 The sera contained virus-neutralizing antibodies of titres ranging between 1 : 4 and 1 : 8, and were positive in a PRV-specific-ELISA only to a serum dilution of 1 : 20 (data not shown). After challenge infection all animals showed severe and progressing clinical signs (fever, respiratory and central nervous symptoms) and were killed between days 5 and 7 after infection.
Discussion
In the present study a quantitative RT–PCR was established and used to analyse the cytokine response, after specific restimulation of lymphocytes, with respect to a Th1- or a Th2-type response and as a possible prediction of efficient immunization. There is increasing evidence that cell-mediated immunity (in particular specific memory T cells) is important for inducing long-lived protective immunity against herpesviruses.11,37 In humans, mice or cattle, protective immunity against various viruses, including herpesviruses, is characterized by a Th1-type immune response according to the induced cytokine pattern.25,28,38–40 In pigs, however, information on the Th1/Th2 bias after viral infection or antiviral vaccination is still limited.
We determined the induction of typical Th1-type (IL-2, IFN-γ) and Th2-type (IL-4, IL-10) cytokines19 in PBMC from pigs, which were solidly protected against PRV challenge infection. After constructing non-homologous competitive DNA fragments (MIMICS), as described by Siebert & Larrick, 1993,41 the RT–qcPCR method was used to determine cytokine induction. This method allowed a rapid and reliable relative quantification of the different cytokine mRNAs using a single cDNA prepared from PBMC. To ensure cDNA integrity and concentration, which might differ between RNA samples, the expression of the housekeeping gene GAPDH was additionally quantified as an endogenous control. No significant differences of GAPDH transcription were found in PBMC after stimulation (with virus or Con A) and in non-stimulated PBMC derived from the different pigs. Phytohemagglutinin (PHA) for lectin-mediated cytokine stimulation in porcine PBMC was recently reported to affect transcription of GAPDH mRNA as well as to stimulate IL-4 in PBMC from a single PRV-infected pig.35 Our results confirm that Con A treatment of PBMC from PRV-immune or -naive pigs is well suited for the reproducible induction of the cytokines tested,34 and demonstrate that GAPDH mRNA can be used as a reliable endogenous control.
PBMC from naive and PRV-primed pigs were stimulated in vitro with PRV to evaluate the cytokine response reflecting innate and antigen recall reactions. The RT–qcPCR results clearly demonstrated that, upon in vitro PRV re-exposure using either live or β-propiolactone-inactivated virus, transcription of IL-2 and IFN-γ was up-regulated in immune cells from all PRV-primed pigs. After restimulation, only PBMC from immune animals displayed a 40- to 120-fold increase in IL-2 and a 30- to 60-fold increase in IFN-γ transcription, surpassing even the ICT after 24-hr of stimulation with Con A (Table 2). Using a different approach of quantitative competitive PCR, Dufour et al.35 also reported an increased transcription of IL-2 and IFN-γ in PRV-restimulated PBMC from a single animal. The increased transcription of Th1-type cytokines in PBMC of immunized pigs was specific for PRV, as addition of the closely related alphaherpesvirus BHV-1 did not result in increased transcription of IL-2 and IFN-γ. Re-exposure with inactivated and live PRV led to a similar stimulation of IL-2 and IFN-γ, which indicates that in vitro PRV replication was not necessary, at least during the 24-hr stimulation protocol. The restimulation of PRV-specific memory cells is probably responsible for this type of cytokine pattern. Recently, we reported that PRV-specific, porcine memory T lymphocytes responded both to PRV and PRV glycoprotein gC-specific peptides, which is accompanied with an increased secretion of IL-2.7 Secretion of IFN-γ has also been found after specific restimulation of PBMC from PRV-primed pigs.29,30
In vitro PRV stimulation did not induce or enhance IL-4 or IL-10 in PBMC from naive or immune animals. IL-4-specific mRNA was not detectable in non-stimulated or PRV-exposed porcine PBMC, although this RT–PCR demonstrated high sensitivity and IL-4 mRNA was found regularly 24 hr after Con A treatment. In no case was a significant down-regulation of IL-10 found after the addition of live or inactivated PRV, as has been recently reported for one naive and one immune pig.35 In the present work a relatively high basal IL-10 transcription was reproducibly found in non-stimulated PBMC from the seven animals investigated. Similar results have been reported by others using either non-competitive34 or competitive RT–PCR,36 not only with porcine PMBC, but also with feline, bovine or simian PBMC.31,42,43 The reason for such a high basal IL-10 transcription in these animals is unclear, and whether IL-10 protein is also expressed and secreted in higher levels than other cytokines remains to be shown. Technical reasons for this result, such as cell culture conditions or the procedure used for blood sampling, cannot be excluded completely, although we found no differences on varying the source of media, serum or culture vessels (data not shown). As IL-10 is able to regulate the activation of Th1 cells,44,45 it can therefore down-regulate a Th1-mediated inflammatory immune response.46 Its immunomodulatory role has also been described in pigs.47 Specific viral restimulation of PBMC, e.g. from HSV-seropositive individuals, induced not only IL-2 and IFN-γ but also showed an enhanced secretion of IL-10.40 Similarly to human IL-10, bovine IL-10 was reported to be expressed in all Th cell subsets and not, as is known for mice, selectively in Th2 cells.23,48 It remains to be clarified whether the unique composition of T-cell subsets of porcine PBMC18 might be responsible for the markedly high constitutive expression of IL-10, which consequently requires identifying the PBMC subpopulation that predominantly produces IL-10.
The results presented in this work indicate that effective in vivo priming of pigs with PRV leads to the generation of PBMC that probably contain PRV-specific memory cells, which are characterized by a Th1-like cytokine pattern upon in vitro recall stimulation. Protective vaccination of mice against PRV generated an antiviral Th1-type immune response27,28 comparable to that of HSV.25,26 A Th1-type cytokine-mediated immune response was crucial for preventing mice from lethal challenge with HSV, whereas a Th2-type cytokine-mediated response led to an increased rate of morbidity and mortality of HSV-infected mice.25 Similarly, protective vaccination of cattle against BHV-1 was improved by administration of the Th1 cytokine, IL-2.49,50
An important role of Th1-type cytokines for protecting pigs against PRV challenge infection is indicated by the results obtained with PBMC from three DNA-vaccinated pigs. The animals were immunized three times with a plasmid encoding the complete glycoprotein, gC, of PRV, but were not protected against challenge infection with the highly virulent PRV strain, NIA-3. Although gC plays an important role in mediating a cellular immune response against PRV,7,14 more recent data showed insufficient protection against lethal challenge after administration of gC alone.2 Using the described RT–qcPCR, no increase in IL-2 and IFN-γ transcription could be measured after in vitro restimulation with PRV of the PBMC derived from the DNA-immunized animals (Table 2). The presence of low, specific serum antibody titres confirm immunoreactivity after gC DNA administration; however, this DNA immunization did not result in the priming of a solid, Th1-type cytokine response. IL-2 and IFN-γ represent pivotal cytokines produced by Th1 cells, which provide help for the generation of virus-neutralizing serum antibodies as well as cytolytic T cells.19 The success of genetic vaccination, e.g. against herpesviruses, seems to be determined by the induction of a Th1-type immune response, which is directed by the nature and type of antigen, route of administration, immune modulatory or adjuvant effect of the plasmid used for immunization, and co-expression of cytokines (reviewed in ref. 50). Determination of the cytokine pattern, e.g. using the simple, but reliable and sensitive, RT–qcPCR procedure used in this work, in PBMC from immunized and non-immunized animals, might be of prognostic value for the induction of protective immunity. More detailed studies will demonstrate whether evaluation of the induced cytokines would be of value for improving the choice and composition of antigen(s), which might be feasible already early at the onset of immunity and independent from the detectable generation of serum antibodies.
Acknowledgments
Part of this study was financially supported by EVAX GmbH, Martinsried-Munich, Germany. DNA purification of plasmid gC-CMV was performed by S. Resch, EVAX GmbH. The authors thank Lothar Stitz (Federal Res. Ctr. Virus Dis. Animals, Tübingen) for valuable discussions and critically commenting on the manuscript.
Glossary
Abbreviations
- BHV
bovine herpesvirus
- Con A
concanavalin A
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- ICT
induction coefficient of transcription
- PBMC
peripheral blood mononuclear cells
- PRV
pseudorabies virus
- RT–qcPCR
quantitative, competitive reverse transcription–polymerase chain reaction
References
- 1.Wittmann G, Rziha H-J. Aujeszky's disease (Pseudorabies) in pigs. In: Wittmann G, editor. Herpesvirus Diseases of Cattle, Horses, and Pigs. Boston, MA: Kluwer Academic Publishers; 1989. pp. 230–325. [Google Scholar]
- 2.Gerdts V, Jöns A, Mettenleiter TC. Potency of an experimental DNA vaccine against Aujeszky's disease in pigs. Vet Microbiol. 1999;66:1–13. doi: 10.1016/s0378-1135(98)00300-9. 10.1016/S0378-1135(98)00300-9. [DOI] [PubMed] [Google Scholar]
- 3.Gerdts V, Jöns A, Makoschey B, Visser N, Mettenleiter TC. Protection of pigs against Aujeszky's disease by DNA vaccination. J Gen Virol. 1997;78:2139–46. doi: 10.1099/0022-1317-78-9-2139. [DOI] [PubMed] [Google Scholar]
- 4.Haagmans BL, van Rooij EM, Dubelaar M, et al. Vaccination of pigs against pseudorabies virus with plasmid DNA encoding glycoprotein D. Vaccine. 1999;17:1264–71. doi: 10.1016/s0264-410x(98)00349-1. 10.1016/S0264-410X(98)00349-1. [DOI] [PubMed] [Google Scholar]
- 5.van Rooij EM, Haagmans BL, de Visser YE, de Bruin MG, Boersma W, Bianchi AT. Effect of vaccination route and composition of DNA vaccine on the induction of protective immunity against pseudorabies infection in pigs. Vet Immunol Immunopathol. 1998;66:113–26. doi: 10.1016/s0165-2427(98)00186-x. 10.1016/S0165-2427(98)00186-X. [DOI] [PubMed] [Google Scholar]
- 6.Zinkernagel RM, Bachmann MF, Kundig TM, Oehen S, Pirchet H, Hengartner H. On immunological memory. Annu Rev Immunol. 1996;14:333–67. doi: 10.1146/annurev.immunol.14.1.333. [DOI] [PubMed] [Google Scholar]
- 7.Ober BT, Summerfield A, Mattlinger C, et al. Vaccine-induced, pseudorabies virus-specific, extrathymic CD4+ CD8+ memory T-helper cells in swine. J Virol. 1998;72:4866–73. doi: 10.1128/jvi.72.6.4866-4873.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawashima K, Platt KB. The effect of human recombinant interleukin 2 on the porcine immune response to a pseudorabies virus subunit vaccine. Vet Immunol Immunopathol. 1989;22:345–53. doi: 10.1016/0165-2427(89)90170-0. [DOI] [PubMed] [Google Scholar]
- 9.Ramshaw IA, Ramsay AJ, Karupiah G, Rolph MS, Mahaligan S, Ruby JC. Cytokines and immunity to viral infections. Immunol Rev. 1997;159:119–35. doi: 10.1111/j.1600-065x.1997.tb01011.x. [DOI] [PubMed] [Google Scholar]
- 10.Nash AA, Cambouropoulos P. The immune response to herpes simplex virus. Semin Virol. 1993;4:181–6. [Google Scholar]
- 11.Manickan E, Rouse BT. Roles of different T-cell subsets in control of herpes simplex virus infection determined by using T-cell-deficient mouse-models. J Virol. 1995;69:8178–9. doi: 10.1128/jvi.69.12.8178-8179.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimman TG, DeBruin TG, Voermans JJ, Bianchi AT. Cell-mediated immunity to pseudorabies virus: cytolytic effector cells with characteristics of lymphokine-activated killer cells lyse virus-infected and glycoprotein gB- and gC-transfected L14 cells. J Gen Virol. 1996;77:987–90. doi: 10.1099/0022-1317-77-5-987. [DOI] [PubMed] [Google Scholar]
- 13.Kimman TG, De Bruin TM, Voermans JJ, Peeters BP, Bianchi AT. Development and antigen specificity of the lymphoproliferation responses of pigs to pseudorabies virus: dichotomy between secondary B- and T-cell responses. Immunology. 1995;86:372–8. [PMC free article] [PubMed] [Google Scholar]
- 14.Zuckermann FA, Zsak L, Mettenleiter TC, Ben Porat T. Pseudorabies virus glycoprotein gIII is a major target antigen for murine and swine virus-specific cytotoxic T lymphocytes. J Virol. 1990;64:802–12. doi: 10.1128/jvi.64.2.802-812.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Summerfield A, Rziha HJ, Saalmüller A. Functional characterization of porcine CD4+ CD8+ extrathymic T lymphocytes. Cell Immunol. 1996;168:291–6. doi: 10.1006/cimm.1996.0078. 10.1006/cimm.1996.0078. [DOI] [PubMed] [Google Scholar]
- 16.Zuckermann FA, Husmann RJ. Functional and phenotypic analysis of porcine peripheral blood CD4/CD8 double-positive T cells. Immunology. 1996;87:500–12. [PMC free article] [PubMed] [Google Scholar]
- 17.Ober BT, Teufel B, Wiesmüller KH, et al. The porcine humoral immune response against pseudorabies virus specifically targets attachment sites on glycoprotein gC. J Virol. 2000;74:1752–60. doi: 10.1128/jvi.74.4.1752-1760.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saalmüller A, Pauly T, Hohlich BJ, Pfaff E. Characterization of porcine T lymphocytes and their immune response against viral antigens. J Biotechnol. 1999;73:223–33. doi: 10.1016/s0168-1656(99)00140-6. 10.1016/S0168-1656(99)00140-6. [DOI] [PubMed] [Google Scholar]
- 19.Abbas AK, Murphy KM, Sher A. Functional diversity of helper T lymphocytes. Nature. 1996;383:787–93. doi: 10.1038/383787a0. [DOI] [PubMed] [Google Scholar]
- 20.Brown WC, Rice-Ficht AC, Estes DM. Bovine type 1 and type 2 responses. Vet Immunol Immunopathol. 1998;63:45–55. doi: 10.1016/s0165-2427(98)00081-6. 10.1016/S0165-2427(98)00081-6. [DOI] [PubMed] [Google Scholar]
- 21.Romagnani S. Lymphokine production by human T cells in disease states. Annu Rev Immunol. 1994;12:227–57. doi: 10.1146/annurev.iy.12.040194.001303. [DOI] [PubMed] [Google Scholar]
- 22.Sinigaglia F, D'ambrosio D, Panina-Bordignon P, Rogge L. Regulation of the IL-12/IL-12R axis: a critical step in T-helper cell differentiation and effector function. Immunol Rev. 1999;170:65–72. doi: 10.1111/j.1600-065x.1999.tb01329.x. [DOI] [PubMed] [Google Scholar]
- 23.Sinigaglia F, D'ambrosio D, Rogge L. Type I interferons and the Th1/Th2 paradigm. Dev Comp Immunol. 1999;23:657–63. doi: 10.1016/s0145-305x(99)00039-7. 10.1016/S0145-305X(99)00039-7. [DOI] [PubMed] [Google Scholar]
- 24.Biron CA. Initial and innate responses to viral infections – pattern setting in immunity or disease. Curr Opin Microbiol. 1999;2:374–81. doi: 10.1016/s1369-5274(99)80066-6. 10.1016/S1369-5274(99)80066-6. [DOI] [PubMed] [Google Scholar]
- 25.Sin JI, Kim JJ, Boyer JD, Ciccarelli RB, Higgins TJ, Weiner DB. In vivo modulation of vaccine-induced immune responses toward a Th1 phenotype increases potency and vaccine effectiveness in a herpes simplex virus type 2 mouse model. J Virol. 1999;73:501–9. doi: 10.1128/jvi.73.1.501-509.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu Z, Manickan E, Rouse BT. Role of interferon-gamma in immunity to herpes simplex virus. J Leukoc Biol. 1996;60:528–32. doi: 10.1002/jlb.60.4.528. [DOI] [PubMed] [Google Scholar]
- 27.Bianchi AT, Moonen-Leusen HW, van Milligen FJ, Savelkoul HF, Zwart RJ, Kimman TG. A mouse model to study immunity against pseudorabies virus infection: significance of CD4+ and CD8+ cells in protective immunity. Vaccine. 1998;16:1550–8. doi: 10.1016/s0264-410x(98)00044-9. 10.1016/S0264-410X(98)00044-9. [DOI] [PubMed] [Google Scholar]
- 28.Schijns VE, Haagmans BL, Horzinek MC. IL-12 stimulates an antiviral type 1 cytokine response but lacks adjuvant activity in IFN-gamma-receptor-deficient mice. J Immunol. 1995;155:2525–32. [PubMed] [Google Scholar]
- 29.Mateu de Antonio E, Husmann RJ, Hansen R, et al. Quantitative detection of porcine interferon-gamma in response to mitogen, superantigen and recall viral antigen. Vet Immunol Immunopathol. 1998;61:265–77. doi: 10.1016/s0165-2427(97)00141-4. 10.1016/S0165-2427(97)00141-4. [DOI] [PubMed] [Google Scholar]
- 30.Zuckermann FA, Husmann RJ, Schwartz R, Brandt J, Mateu de Antonio E, Martin S. Interleukin-12 enhances the virus-specific interferon gamma response of pigs to an inactivated pseudorabies virus vaccine. Vet Immunol Immunopathol. 1998;63:57–67. doi: 10.1016/s0165-2427(98)00082-8. 10.1016/S0165-2427(98)00082-8. [DOI] [PubMed] [Google Scholar]
- 31.Rottman JB, Tompkins WA, Tompkins MB. A reverse transcription-quantitative competitive polymerase chain reaction (RT–qcPCR) technique to measure cytokine gene expression in domestic mammals. Vet Pathol. 1996;33:242–8. doi: 10.1177/030098589603300217. [DOI] [PubMed] [Google Scholar]
- 32.Zimmermann K, Mannhalter JW. Technical aspects of quantitative competitive PCR. Biotechniques. 1996;21:268–9. doi: 10.2144/96212rv01. [DOI] [PubMed] [Google Scholar]
- 33.Mansfield LS, Urban JF, Holley-Shanks RR, et al. Construction of internal cDNA competitors for measuring IL-10 and IL-12 cytokine gene expression in swine. Vet Immunol Immunopathol. 1998;65:63–74. doi: 10.1016/s0165-2427(98)00106-8. 10.1016/S0165-2427(98)00106-8. [DOI] [PubMed] [Google Scholar]
- 34.Dozois CM, Oswald E, Gautier N, Serthelon JP, Fairbrother JM, Oswald IP. A reverse transcription-polymerase chain reaction method to analyze porcine cytokine gene expression. Vet Immunol Immunopathol. 1997;58:287–300. doi: 10.1016/s0165-2427(97)00039-1. 10.1016/S0165-2427(97)00039-1. [DOI] [PubMed] [Google Scholar]
- 35.Dufour V, Arnauld C, Lantz O, et al. Quantification of porcine cytokine gene expression using RT-PCR, a homologous internal control and chemiluminescence for microplate detection. J Immunol Methods. 1999;229:49–60. doi: 10.1016/s0022-1759(99)00105-2. 10.1016/S0022-1759(99)00105-2. [DOI] [PubMed] [Google Scholar]
- 36.Reddy NR, Wilkie BN. Quantification of porcine cytokine and beta 2-microglobulin mRNA expression by reverse transcription polymerase chain reaction. J Immunol Methods. 2000;233:83–93. doi: 10.1016/s0022-1759(99)00188-x. 10.1016/S0022-1759(99)00188-X. [DOI] [PubMed] [Google Scholar]
- 37.Manickan E, Rouse RJ, Yu Z, Wire WS, Rouse BT. Genetic immunization against herpes simplex virus. Protection is mediated by CD4+ T lymphocytes. J Immunol. 1995;155:259–65. [PubMed] [Google Scholar]
- 38.Romagnani S. The Th1/Th2 paradigm. Immunol Today. 1997;18:263–6. doi: 10.1016/s0167-5699(97)80019-9. 10.1016/S0167-5699(97)01070-0. [DOI] [PubMed] [Google Scholar]
- 39.Hughes HP, Campos M, Drunen Littel-van den Hurk S, et al. Multiple administration with interleukin-2 potentiates antigen-specific responses to subunit vaccination with bovine herpesvirus-1 glycoprotein IV. Vaccine. 1992;10:226–30. doi: 10.1016/0264-410x(92)90157-f. [DOI] [PubMed] [Google Scholar]
- 40.Carmack MA, Yasukawa LL, Chang SY, et al. T cell recognition and cytokine production elicited by common and type-specific glycoproteins of herpes simplex virus type 1 and type 2. J Infect Dis. 1996;174:899–906. doi: 10.1093/infdis/174.5.899. [DOI] [PubMed] [Google Scholar]
- 41.Siebert PD, Larrick JW. PCR MIMICS competitive DNA fragments for use as internal standards in quantitative PCR. Biotechniques. 1993;14:244–9. [PubMed] [Google Scholar]
- 42.Benveniste O, Vaslin B, Villinger F, Le Grand R, Ansari AA, Dormont D. Cytokine mRNA levels in unmanipulated (ex vivo) and in vitro stimulated monkey PBMCs using a semi-quantitative RT–PCR and high sensitivity fluorescence-based detection strategy. Cytokine. 1996;8:32–41. doi: 10.1006/cyto.1996.0005. 10.1006/cyto.1996.0005. [DOI] [PubMed] [Google Scholar]
- 43.Covert J, Splitter G. Detection of cytokine transcriptional profiles from bovine peripheral blood mononuclear cells and CD4+ lymphocytes by reverse transcriptase polymerase chain reaction. Vet Immunol Immunopathol. 1995;49:39–50. doi: 10.1016/0165-2427(95)05451-b. 10.1016/0165-2427(95)05451-B. [DOI] [PubMed] [Google Scholar]
- 44.Fiorentino DF, Zlotnik A, Mosmann TR, Howard M, O'garra A. IL-10 inhibits cytokine production by activated macrophages. J Immunol. 1991;147:3815–22. [PubMed] [Google Scholar]
- 45.Fiorentino DF, Bond MW, Mosmann TR. Two types of mouse T helper cell. IV. Th2 clones secrete a factor that inhibits cytokine production by Th1 clones. J Exp Med. 1989;170:2081–95. doi: 10.1084/jem.170.6.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gazzinelli RT, Amichay D, Sharton-Kersten T, Grunwald E, Farber JM, Sher A. Role of macrophage-derived cytokines in the induction and regulation of cell-mediated immunity to Toxoplasma gondii. Curr Top Microbiol Immunol. 1996;219:127–39. doi: 10.1007/978-3-642-51014-4_12. [DOI] [PubMed] [Google Scholar]
- 47.Blancho G, Gianello P, Germana S, Baetscher M, Sachs DH, LeGuern C. Molecular identification of porcine interleukin 10: regulation of expression in a kidney allograft model. Proc Natl Acad Sci USA. 1995;92:2800–4. doi: 10.1073/pnas.92.7.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brown WC, Woods VM, Chitko-McKown CG, Hash SM, Rice-Ficht AC. Interleukin-10 is expressed by bovine type 1 helper, type 2 helper, and unrestricted parasite-specific T-cell clones and inhibits proliferation of all three subsets in an accessory-dependent manner. J Virol. 1994;62:4697–708. doi: 10.1128/iai.62.11.4697-4708.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Reddy DN, Reddy PG, Xue W, Minocha HC, Daley MJ, Blecha F. Immunopotentiation of bovine respiratory disease virus vaccines by interleukin-1 beta and interleukin-2. Vet Immunol Immunopathol. 1993;37:25–38. doi: 10.1016/0165-2427(93)90013-t. [DOI] [PubMed] [Google Scholar]
- 50.Davis HL, McCluskie MJ. DNA vaccines for viral diseases. Microbes Infect. 1999;1:7–21. doi: 10.1016/s1286-4579(99)80009-4. 10.1016/S1286-4579(99)80009-4. [DOI] [PubMed] [Google Scholar]