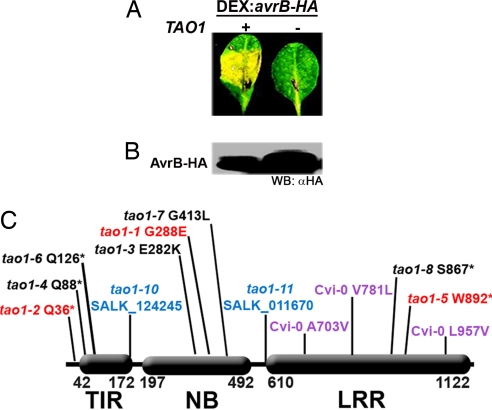
Fig. 1.
TAO1 is a TIR-NB-LRR R protein required for AvrB-induced chlorosis in rpm1 host plants. (A) Mt-0 leaves were inoculated with Agrobacterium containing T-DNA with a DEX:avrB-HA transgene (4). Leaves were treated with DEX 48 h after inoculation. The picture was taken 72 h after inoculation. (B) Western blot showing accumulation of AvrB-HA at 8 h after DEX treatment. (C) Deduced structure of TAO1 alleles recovered in Mt-0 (red and black), Col-0 Salk T-DNA insertion lines (blue), and as polymorphisms in Cvi-0 (purple). For all missense mutations, the wild-type Mt-0 residue is listed first. TIR, amino acids 42–172. NB, amino acids 197–492. LRR, amino acids 610-1122. All TAO1 alleles in Mt-0 were generated by EMS mutagenesis except tao1–8, which is a fast neutron deletion of 1 bp in codon S667. Red tao1 alleles represent alleles that were out-crossed away from the DEX:avrB-HA transgene and used for further analyses. The tao1–10 (Salk_124245) insertion begins at amino acid 168. The tao1–11 (Salk_011670) insertion begins at amino acid 597. The only amino acid difference between Mt-0 and Col-0 is V489M. Genomic TAO1 sequences for Mt-0 (EU031442) and Cvi-0 (EU031443) have been deposited in GenBank.