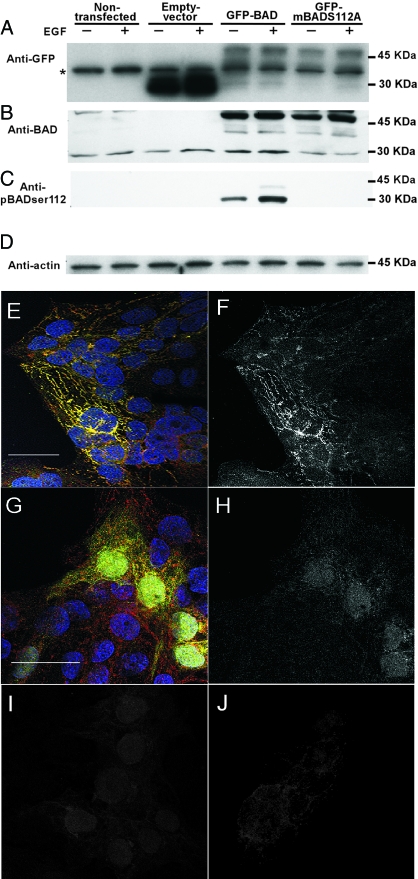
Figure 3.
The expression and interaction of BAD and GFP-BAD in transfected JEG3 cells. Lysates of JEG3 cells stably transfected with vector alone, GFP-BAD, or GFP-mBADS112A were screened to identify GFP (A), endogenous BAD (23 kDa; B), GFP-BAD (46 kDa; B), phosphorylated BADser112 (pBAD-ser112; C), and actin (D). An asterisk (A) indicates a nonspecific band present in all paradigms using the anti-GFP antibody. The level of phosphorylated BADser112 was below the level of detection in the JEG3 and vector control cells examined in the blot of C. E, A confocal fluorescence microscopy image of JEG3 cells transfected with GFP-BAD and immunostained for both endogenous BAD (anti-BAD primary antibody with secondary antibody conjugated to Alexa 546, red fluorescence) and GFP-BAD fusion protein (anti-GFP primary antibody with secondary antibody conjugated to Alexa 488, green fluorescence). A punctate yellow signal resulted in transfected cells, consistent with preferential localization in mitochondria. This yellow signal resulted from the spectral overlap of the red and green fluorescence, indicating endogenous BAD and the GFP-BAD fusion protein were in the same subcellular compartment. A FRET signal was also detected in this field (F) to confirm that there was not only colocalization, but also a molecular interaction, of endogenous BAD and the GFP-BAD fusion protein. Control images of JEG3 cells transfected with GFP-only vector (G) showed punctate red and diffuse green fluorescence throughout transfected cells, with no punctate yellow signal, indicating no specific subcellular colocalization. There was no specific FRET signal detected in these cultures (H). Controls for the FRET analysis are shown for the spectral overlap of the fluorophores for the anti-BAD labeling alone (I) and anti-GFP labeling alone (J). This spectral overlap fluorescence was subtracted from the FRET-calculated images as shown in F and H (bar, 10 μm).