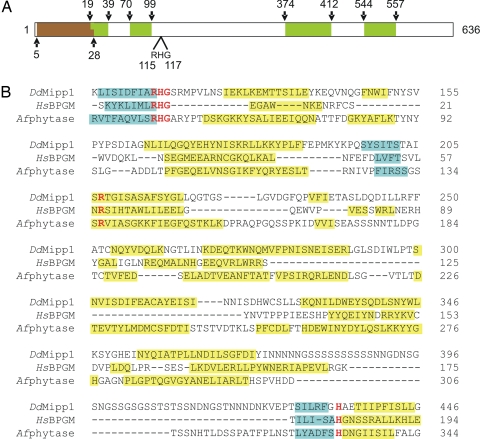
Fig. 1.
Alignment of DdMipp1, HsBPGM, and Afphytase. (A) Schematic of DdMipp1. The predicted transmembrane domain (according to Peptool, v2) is shown in brown; Ser/Asn-rich, simple-sequence repeats are shown in green. (B) D. discoideum (Dd) Mipp1A (DDB0186447; www.dictybase.org), human (Hs) BPGM (PDB 1T8P), and Aspergillus ficuum (Af) phytase (PDB 1IHP). Catalytically important residues (26, 40, 41) that are conserved in all three proteins are shown in red type. The PHYRE server provided the predicted secondary structural elements of DdMipp1 and the actual secondary structures of BPGM and phytase (blue, beta-sheet; yellow, helix; the graphic does not distinguish 310 helices from alpha-helices).