Abstract
The complete absence of dystrophin causes Duchenne muscular dystrophy. Its restoration by greater than 20% is needed to reduce muscle pathology and improve muscle force. Dystrophin levels lower than 20% are considered therapeutically irrelevant but are associated with a less severe phenotype in certain Becker muscular dystrophy patients. To understand the role of low-level dystrophin expression, we compared muscle force and pathology in mdx3cv and mdx4cv mice. Dystrophin was eliminated in mdx4cv mouse muscle but was expressed in mdx3cv mice as a near full-length protein at ∼5% of normal levels. Consistent with previous reports, we found dystrophic muscle pathology in both mouse strains. Surprisingly, mdx3cv extensor digitorium longus muscle showed significantly higher tetanic force and was also more resistant to eccentric contraction-induced injury than mdx4cv extensor digitorium longus muscle. Furthermore, mdx3cv mice had stronger forelimb grip strength than mdx4cv mice. Immunostaining revealed utrophin up-regulation in both mouse strains. The dystrophin-associated glycoprotein complex was also restored in the sarcolemma in both strains although at levels lower than those in normal mice. Our results suggest that subtherapeutic expression levels of near full-length, membrane-bound dystrophin, possibly in conjunction with up-regulated utrophin levels, may help maintain minimal muscle force but not arrest muscle degeneration or necrosis. Our findings provide valuable insight toward understanding delayed clinical onset and/or slow disease progression in certain Becker muscular dystrophy patients.
Duchenne muscular dystrophy (DMD) results from mutations in the dystrophin gene.1 In the striated muscle of DMD patients, dystrophin is essentially eliminated. Dystrophin is an important cytoskeleton protein. It protects the sarcolemma from the shearing stress generated during muscle contraction. In the absence of dystrophin, sarcolemma integrity is compromised. This sensitizes myofibers to contraction-induced injury. As a result, muscle cells undergo degeneration and necrosis. Eventually, muscle is replaced by adipose and fibrous tissues and loses contractility. DMD patients experience difficulties in moving and/or climbing at 3 to 5 years of age. Thereafter, clinical progression follows a catastrophic downhill track. Patients are confined to a wheelchair at ∼11 years of age and die prematurely before age 30.2
Dystrophin gene mutation also causes Becker muscular dystrophy (BMD), a milder allelic form. In BMD patients, symptoms usually start in the teenage years and progress slowly. In contrast to DMD, dystrophin expression is not lost in BMD patients.3 In general, it falls into two categories. Some BMD patients express an internally truncated but partially functional dystrophin. Some of these abbreviated dystrophin isoforms can be quite effective in halting disease progression.4,5 In fact, several gene therapy strategies are based on expressing internally deleted dystrophin. Among these, microdystrophin, minidystrophin, and exon-skipping approaches have shown great promise in ameliorating disease in animal models and are currently in the early phase of clinical trials.
In other BMD patients, dystrophin expression is reduced. In these patients, there seems a clear correlation between the amount of dystrophin protein and the clinical phenotype. Patients who have ≥20% of the normal dystrophin levels usually display mild disease.6,7,8 Most of them are ambulant beyond age 20.6,7,8 Patients who have less than 20% of the levels show an intermediate phenotype between typical DMD and BMD.6,9,10 They become wheelchair-bound between 14 and 20 years of age, several years later than DMD patients.6
Investigators have also studied the effect of varying dystrophin levels in mdx mice, a mouse model for DMD.11,12,13 Uniform expression of full-length dystrophin or minidystrophin at 20% or higher levels results in remarkable improvement in muscle pathology and strength in transgenic mdx mice.12,13 Muscle degeneration/regeneration, sarcolemma leakage, and muscle-specific force are all normalized. Few studies have evaluated the effect of low level (<20%) dystrophin expression in animal models of DMD. Wells and colleagues11 reported partial amelioration of muscle disease in transgenic mdx mice that express a minidystrophin gene at ∼17% of the normal level. They observed a significant reduction in muscle degeneration but the serum creatine kinase (CK) level remained high.11
To better understand the effect of subtherapeutic level dystrophin expression in mice, we compared muscle pathology and force in the limb muscles of BL6, mdx3cv, and mdx4cv mice. Mdx3cv and mdx4cv mice are N-ethylnitrosourea (ENU)-induced mouse models for DMD.14 They are on the BL6 background. Different point mutations in the dystrophin gene disrupt normal dystrophin expression in these mice.15,16 Mdx4cv muscle represents a true dystrophin-null model. However, mdx3cv muscle expresses a low-level near full-length dystrophin protein.15,16,17,18,19 Consistent with previous reports, we found characteristic muscle pathology in both mdx3cv and mdx4cv mice.15,17 Surprisingly, the extensor digitorium longus (EDL) muscle strength was significantly preserved in mdx3cv mice albeit at a level lower than that of normal mice. The mdx3cv EDL muscle was also partially protected from eccentric contraction-induced injury. Furthermore, mdx3cv mice performed better than mdx4cv mice on a forelimb grip strength assay. Additional studies showed utrophin up-regulation in both mdx3cv and mdx4cv muscles. The dystrophin-associated glycoprotein complex (DGC) was also weakly restored in both strains. Using antibodies against different regions of dystrophin, we found uniform low-level expression of a membrane-bound dystrophin protein in mdx3cv, but not mdx4cv, muscles. Taken together, our results provide the first evidence that subtherapeutic levels of a membrane-bound dystrophin expression may help preserve minimal muscle force. However, this low-level dystrophin expression is insufficient to prevent sarcolemma injury neither can it stop myofiber degeneration.
Materials and Methods
Animals
All animal experiments were approved by the Animal Care and Use Committee of the University of Missouri and were in accordance with National Institutes of Health guidelines. BL6 (C57BL/6J), mdx3cv (B6Ros.Cg-Dmdmdx-3Cv/J), and mdx4cv (B6Ros.Cg-Dmdmdx-4Cv/J) mice were purchased from The Jackson Laboratory (Bar Harbor, ME). Male mice (3, 6, and 12 months of age) were used in this study. All experimental mice were housed in a specific pathogen-free facility and were kept under a 12-hour light (25 lux)/12-hour dark cycle with free access to food and water.
Histopathology Studies
Hematoxylin and eosin (H&E) staining was used to reveal general histology and centrally nucleated myofibers. Macrophage infiltration in muscle was evaluated by nonspecific esterase (α-naphthyl butyrate esterase) staining according to a published protocol.20,21 Macrophages are dark brown cells scatted between myofibers. Sarcolemma integrity was determined with an Evans blue dye (EBD) uptake assay described previously.20 Briefly, EBD (10 mg/ml in phosphate-buffered saline, 20 μl/g body weight) was injected into tail vein at 24 hours before muscle harvesting. At 2 hours before tissue collection, mice were run on a 15° downhill treadmill at the speed of 15 m/minute for 30 minutes. Freshly dissected muscle was snap-frozen in liquid nitrogen-cooled isopentane in optimal cutting temperature compound (Sakura Finetek Inc., Torrance, CA). Ten-μm cryosections were visualized under the Texas Red channel with an E800 fluorescence microscope (Nikon, Melville, NY). Nuclei were visualized with 4,6-diamidino-2-phenylindole staining (SlowFade light antifade kit with 4,6-diamidino-2-phenylindole, catalog no. S-24636; Molecular Probes, Eugene, OR).
Immunostaining
Dystrophin was examined with seven epitope-specific antibodies including a mouse monoclonal antibody against the N-terminal domain (Manex1A, 1:300, clone 4C7, IgG2b; a gift from Dr. Glenn Morris, The Robert Jones and Agnes Hunt Orthopaedic Hospital, Shropshire, UK),22 a rabbit polyclonal antibody against spectrin-like repeats 4 to 6 (1:400; Santa Cruz Biotechnology, Santa Cruz, CA), a mouse monoclonal antibody against spectrin-like repeat 11 (Mandys8, 1:200, IgG2b; Sigma, St. Louis, MO), a mouse monoclonal antibody against spectrin-like repeats 14 to 18 (Mandys105, 1:500, clone 8A4, IgG1; a gift from Dr. Glenn Morris),22 a mouse monoclonal antibody against spectrin-like repeat 19/hinge 3 (Manex50, 1:2000, clone 6A9, IgG1; a gift from Dr. Glenn Morris),22 a mouse monoclonal antibody against the C-terminal domain (Mandra1, 1:500, clone 7A10, IgG1; a gift from Dr. Glenn Morris),22 and another mouse monoclonal antibody against the C-terminal domain (Dys-2, 1:30, clone Dy8/6C5, IgG1; Novocastra, Newcastle, UK). Utrophin was examined with a mouse monoclonal antibody against the utrophin N-terminal domain (VP-U579, 1:20, clone DRP3/20C5, IgG1; Vector Laboratories, Burlingame, CA). β-Dystroglycan was revealed with a mouse monoclonal antibody against the C terminus (NCL-b-DG, 1:50, clone 43DAG1/8D5, IgG2a; Novocastra). β-Sarcoglycan was revealed with a mouse monoclonal antibody (NCL-b-SARC, 1:50, clone 5B1, IgG1; Novocastra). Dystrobrevin was revealed with a mouse monoclonal antibody (no. 610766, 1:200, clone 23, IgG1; BD Biosciences, San Diego, CA). Syntrophin was revealed with a pan-syntrophin mouse monoclonal antibody that recognized the PDZ domain (ab11425, 1:200, clone 1351, IgG1; Abcam, Cambridge, MA). Immunostaining was performed essentially as we described before.20,23 To determine the relative immunofluorescence intensity in samples stained with anti-dystrophin or anti-utrophin antibodies, digitized images were quantified using the Image J software (version 1.36b; National Institutes of Health, Bethesda, MD). Three to five random fields were analyzed for each muscle section. At least three different sections were studied for each muscle sample.
Western Blot
Whole muscle lysate was prepared from freshly isolated tibialis anterior (TA) muscle. Briefly, muscle tissue was homogenized in a liquid nitrogen-cooled mortar in a buffer containing 10% sodium dodecyl sulfate, 5 mmol/L ethylenediaminetetraacetic acid, 62.5 mmol/L Tris, pH 6.8, and 1% protease inhibitor (Roche, Indianapolis, IN). After a 2-minute boiling, the homogenate was spun at 14,000 rpm for 2 minutes (Eppendorf centrifuge, model 5417C; Eppendorf-Netheler-Hinz GmbH, Hamburg, Germany). Supernatant was used for Western blot. Protein concentration was determined using a DC protein assay kit (Bio-Rad, Hercules, CA). Muscle lysate (180 μg) was loaded on a 6% sodium dodecyl sulfate-polyacrylamide gel. After electrophoresis, protein was transferred to a polyvinylidene difluoride membrane. Dystrophin was detected with the Mandra1 antibody (1:100, clone 7A10, IgG1; a gift from Dr. Glenn Morris).22 Utrophin was detected with a mouse monoclonal antibody against utrophin amino acid residues 768 to 874 (no. 610896, 1:250, clone 55, IgG1; BD Biosciences). As a loading control, membrane was also probed with an anti-α-tubulin antibody (1:3000, clone B-5-1-2; Sigma).
EDL Muscle Force Measurements
EDL contractile properties were examined using a previously described protocol.20,21,24 The freshly isolated EDL muscle was vertically mounted in a 30°C jacket organ bath containing oxygenated Ringer’s solution (95% O2, 5% CO2). Muscle tendons were attached to a 300B dual-mode servomotor transducer (Aurora Scientific, Inc., Aurora, Canada) with sutures. Optimal muscle length (Lo) was determined as the muscle length at which the maximal twitch force was elicited. At first, all of the myofibers were activated with three 500-ms tetanic stimulations at 150 Hz. After 2 minutes of resting, the absolute twitch force was measured. Muscle force-frequency relationship was then determined with tetanic stimulation at 50, 80, 120, and 150 Hz, respectively. Muscle cross-sectional area was calculated according to the following equation, cross-sectional area = (muscle mass)/(0.44 × Lo × muscle density), where 0.44 represents the ratio of fiber length to optimal muscle length (Lf/Lo) for the EDL muscle. Muscle density is 1.06 g/cm3. The specific force (kN/m2) was calculated by normalizing the absolute muscle force with the cross-sectional area. After tetanic force measurement, the EDL muscle was subjected to an eccentric injury protocol. Briefly, the EDL muscle was stimulated at 150 Hz for 700 ms and an eccentric injury was applied during the last 200 ms by lengthening the muscle by 10% Lo at the speed of 0.5 Lo/second. A total of 10 cycles of eccentric contraction was applied. The maximal isometric tetanic force developed during the first 500 ms of stimulation of the first cycle was designated as 100% force (baseline force). The percentage of tetanic force drop after each eccentric injury was graphed to reflect the level of resistance against contraction-induced injury. Raw data were acquired and analyzed with DMC/DMA software (Version 3.12, Aurora Scientific).
Forelimb Grip Strength Measurement
Forelimb grip strength was measured with a computerized grip strength meter (Columbus Instruments, Columbus, OH). Five sets of measurements were performed in each mouse throughout a period of 60 minutes with a 10-minute rest between sets. In each set of measurements, the mouse was allowed three attempts. The highest force from the three attempts was recorded as the force score for the set. The average value from three highest sets was defined as the forelimb grip strength for the mouse.
Serum CK Assay
Fresh serum was collected by retro-orbital bleeding. The CK level was determined with a CK liqui-UV test kit from Stanbio Laboratory (Boerne, TX).
Statistical Analysis
Data are presented as mean ± SE of mean. Statistical analysis was performed with the SPSS software (SPSS, Chicago, IL). Statistical significance for multiple group comparison was determined by one-way analysis of variance followed by Bonferroni post hoc analysis. Statistical significance for two-group comparison was determined by t-test. Difference was considered significant when P < 0.05.
Results
The Mdx3cv EDL Muscle Showed Significantly Higher Tetanic Force and Was More Resistant to Eccentric Contraction-Induced Injury than the Mdx4cv EDL Muscle
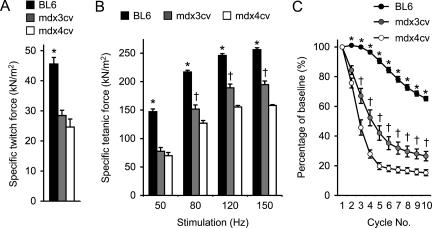
Contractile property of the mdx3cv EDL muscle has not been studied before. We first compared the twitch and tetanic forces in 6-month-old mice. We used age- and sex-matched BL6 mice as the normal control. As expected, the BL6 EDL muscle displayed the highest specific forces under all stimulation conditions. They were significantly higher than those from mdx3cv and mdx4cv muscles (Figure 1). When single twitch and low frequency (50 Hz) tetanic stimulations were applied, mdx3cv and mdx4cv muscles showed similar responses. Surprisingly, when muscles were stimulated at higher stimulation frequencies (80 Hz, 120 Hz, and 150 Hz), the mdx3cv EDL muscle generated significantly stronger force than that of the mdx4cv EDL muscle (Figure 1B).
Figure 1.
Characterization of the EDL muscle contractile profiles in 6-month-old BL6, mdx3cv, and mdx4cv mice. A: Specific twitch force. B: Specific tetanic forces under different stimulation frequencies. C: Muscle force change after 10 rounds of eccentric contraction. Sample size, n = 7 for BL6, n = 8 for mdx3cv, n = 5 for mdx4cv. Asterisk, values in BL6 are significantly different from those in mdx3cv and mdx4cv mice; dagger, values in mdx3cv are significantly different from those in BL6 and mdx4cv mice.
A pivotal feature in DMD is contraction-induced muscle injury. We next examined whether mdx3cv muscle could be protected from eccentric contraction damage. In eccentric contraction, a muscle is stretched beyond its optimal length while the muscle is at its peak tetanic force. This is considered one of the most sensitive physiology assays in evaluating disease progression and therapeutic intervention in muscular dystrophy research. A better force preservation suggests good muscle protection. We applied 10 repeated cycles of eccentric contraction. The BL6 EDL muscle showed moderate force decline through the entire experimental protocol (Figure 1C). Muscle force rapidly dropped to less than 20% of the starting level in the mdx4cv EDL muscle. The mdx3cv EDL muscle also showed a quick drop but to a significantly less extent than that of the mdx4cv EDL muscle (Figure 1C).
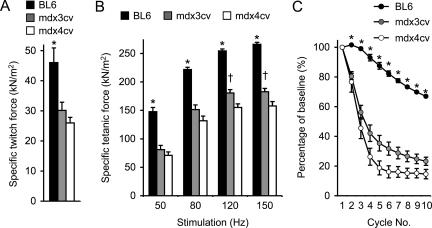
To further extend our observation, we examined muscle contraction in 12-month-old mice (Figure 2). A similar but weaker profile was obtained for the older mdx3cv EDL muscles. Their specific forces were significantly better than those of the mdx4cv EDL muscles at 120-Hz and 150-Hz stimulation frequencies (Figure 2B). In the eccentric contraction assay, they also showed a trend of better force preservation although it did not reach statistical significance (Figure 2C). Despite the difference in the tetanic force and eccentric contraction profiles between mdx3cv and mdx4cv mice, the anatomical features of the EDL muscles were identical between the two groups (Table 1). Both showed significant hypertrophy as demonstrated by the increased EDL muscle weight and the enlarged cross-sectional area (Table 1).
Figure 2.
The EDL muscles of 12-month-old mdx3cv mice display better specific tetanic force than that of age-matched mdx4cv mice at high stimulation frequencies. A: Specific twitch force. B: The force-frequency relationship. C: Relative force decline after 10 rounds of eccentric contraction-induced injury. Sample size, n = 4 for BL6, n = 8 for mdx3cv, n = 6 for mdx4cv. Asterisk, values in BL6 are significantly different from those in mdx3cv and mdx4cv mice; dagger, values in mdx3cv are significantly different from those in BL6 and mdx4cv mice.
Table 1.
Characterization of the EDL Muscle
| Strain | Age (months) | n | Weight (mg) | Lo (mm) | CSA (mm2) |
|---|---|---|---|---|---|
| BL6 | 6 | 8 | 11.20 ± 0.44* | 13.20 ± 0.05 | 1.82 ± 0.07* |
| mdx3cv | 6 | 8 | 18.70 ± 0.40 | 13.80 ± 0.12 | 2.71 ± 0.13 |
| mdx4cv | 6 | 5 | 18.98 ± 0.12 | 13.68 ± 0.13 | 2.98 ± 0.02 |
| BL6 | 12 | 4 | 11.00 ± 0.26* | 12.80 ± 0.12 | 1.84 ± 0.03* |
| mdx3cv | 12 | 10 | 17.76 ± 1.12 | 14.44 ± 0.16 | 2.63 ± 0.14 |
| mdx4cv | 12 | 11 | 18.40 ± 0.36 | 13.76 ± 0.17 | 2.87 ± 0.06 |
Significantly different from those of age-matched mdx3cv and mdx4cv.
Mdx3cv Mice Demonstrated Stronger Forelimb Grip Strength
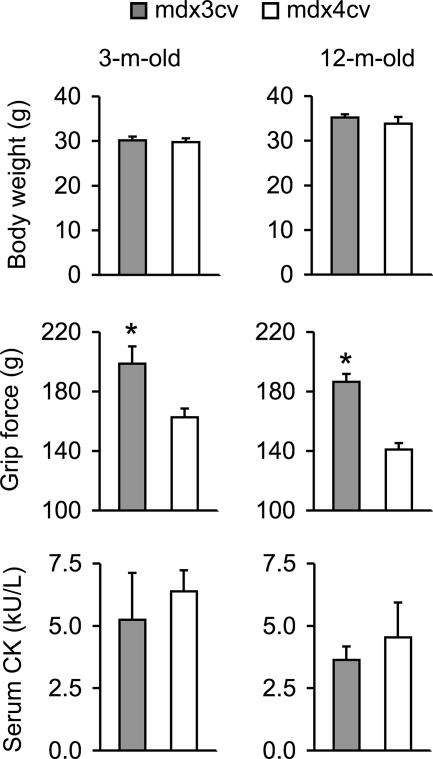
To confirm in vitro force measurement results from the isolated EDL muscles, we compared forelimb grip strength in conscious mdx3cv and mdx4cv mice. Despite the similar body weights, mdx3cv mice showed significantly higher grip force at 3 months and 12 months of age (Figure 3).
Figure 3.
Body weight, grip strength, and the CK levels in 3-month-old and 12-month-old mdx3cv and mdx4cv mice. Sample size for body weight study and grip strength measurement, n = 5 for 3-month-old mdx3cv; n = 15 for 3-month-old mdx4cv; n = 8 for 12-month-old mdx3cv; n = 7 for 12-month-old mdx4cv. Sample size for the CK assay, n = 5 for 3-month-old mdx3cv; n = 14 for 3-month-old mdx4cv; n = 13 for 12-month-old mdx3cv; n = 6 for 12-month-old mdx4cv. Asterisk, values are significantly different from the other group.
Mdx3cv and Mdx4cv Skeletal Muscle Displayed Similar Muscle Pathology
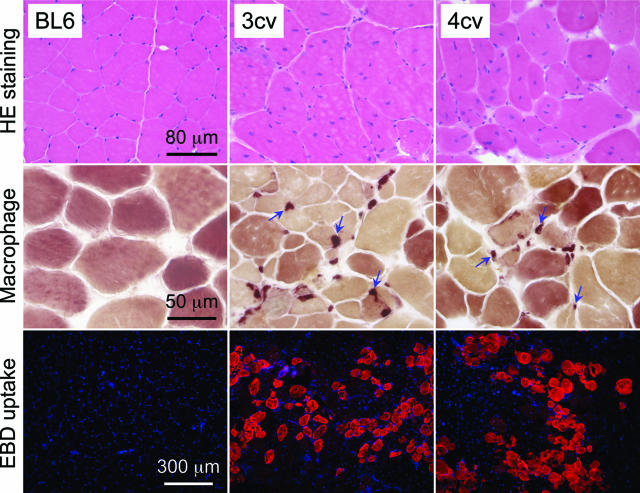
We evaluated muscle morphology by H&E staining. BL6 myofiber had uniform size and peripherally localized nuclei. In both mdx3cv and mdx4cv muscles, there was considerable difference in myofiber size. Some myofibers were several-fold larger than the surrounding myofibers. Furthermore, nuclei were relocated to the center in the majority of the myofibers in mdx3cv and mdx4cv muscles (Figure 4). On average, 84.8 ± 3.2% and 86.7 ± 2.2% myofibers were centrally nucleated in mdx3cv and mdx4cv muscles, respectively.
Figure 4.
Characterization of muscle pathology in BL6, mdx3cv, and mdx4cv mice. Top and middle: Representative photomicrographs of H&E and macrophage (nonspecific esterase) staining in the TA muscles of 6-month-old mice. Blue arrows, macrophages. Bottom: Representative photomicrographs of EBD uptake in the gastrocnemius muscle in 6-month-old mice. The red cytosolic staining marks EBD accumulation in damaged myofibers. Nuclei were stained with 4,6-diamidino-2-phenylindole in blue color.
To study muscle inflammation, we performed nonspecific esterase staining to detect macrophage invasion. We observed macrophage infiltration in the interstitial regions in both mdx3cv and mdx4cv muscles but not in BL6 muscle (Figure 4). The loss of dystrophin weakens sarcolemma strength. This can be documented by the accumulation of EBD inside myofiber. We found extensive EBD uptake in both mdx3cv and mdx4cv muscles but not in BL6 muscle (Figure 4). The serum CK level indirectly reflects sarcolemma damage in all body muscles. Consistent with our EBD uptake results, we found similar levels of CK elevation in mdx3cv and mdx4cv mice (Figure 3).
Representative muscle pathology in the TA muscle and the gastrocnemius muscle is presented in Figure 4 for 6-month-old mice. Similar pathology was observed in 12-month-old mice in other limb muscles and the diaphragm (data not shown). In summary, we found characteristic dystrophic pathology in both mdx3cv and mdx4cv skeletal muscles.
Dystrophin and the DGC Expression in Mdx3cv and Mdx4cv Muscles
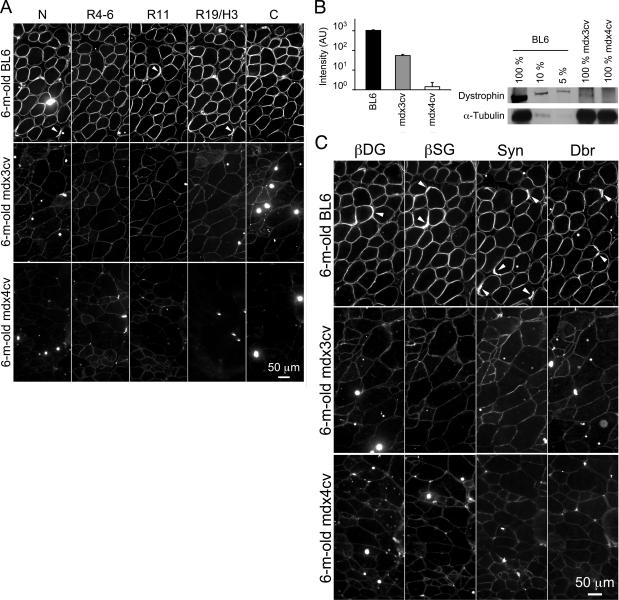
To determine dystrophin expression on the sarcolemma, we performed a comprehensive immunofluorescence staining with a panel of seven epitope-specific antibodies in the EDL, TA, and gastrocnemius muscles, the diaphragm, and the heart. These include the N-terminal-specific manex1A antibody, a repeats 4 to 6-specific polyclonal antibody, the repeat 11-specific mandys8 antibody, the repeats 14 to 18-specific mandys105 antibody, the repeat 19/hinge 3-specific manex50 antibody, and two C-terminal-specific antibodies (dys-2 and mandra1). Figure 5A shows representative staining results from the TA muscle in 6-month-old mice (data not shown for mandys105 and mandra1) (Representative results from the heart were shown in Supplementary Figure S1 at http://ajp.amjpathol.org). Strong immunostaining was observed at the sarcolemma in BL6 muscle. Signals were enriched at the neuromuscular junctions (Figure 5A). A weak but uniform staining was seen at the mdx3cv sarcolemma by all antibodies. No signal was detected with the N-terminal, repeat 19/hinge 3 and C-terminal-specific antibodies on the mdx4cv sarcolemma. However, a faint staining was seen in mdx4cv muscle using repeats 4 to 6 and repeat 11-specific antibodies. These signals seem to be authentic to dystrophin rather than an artificial effect. Using the same antibodies, we did not see any staining in mdx muscle (data not shown).
Figure 5.
Differential expression of dystrophin and the DGC in skeletal muscle of 6-month-old BL6, mdx3cv, and mdx4cv mice. A: Representative photomicrographs of immunofluorescence staining with epitope-specific antibodies that recognize different regions of dystrophin. N, dystrophin N-terminal domain; R4-6, dystrophin rod domain spectrin-like repeats 4 to 6; R11, dystrophin rod domain spectrin-like repeat 11; R19/H3, dystrophin rod domain spectrin-like repeats 19 and hinge 3; C, dystrophin C-terminal domain. B: Left, relative fluorescence signal intensity. AU, artificial unit. n = 15 for BL6, n = 12 for mdx3cv, and n = 9 for mdx4cv. Right, Representative Western blot with an anti-dystrophin C-terminal antibody. C: Representative photomicrographs of immunofluorescence staining with antibodies against selective components of the DGC. βDG, β-dystroglycan; βSG, β-sarcoglycan; Syn, syntrophin; Dbr, dystrobrevin. Arrowhead, neuromuscular junction.
To determine the relative levels of sarcolemmal dystrophin in mdx3cv mice, we quantified the signal intensity in immunofluorescence images. On average, mdx3cv muscle expressed 5.21% of the normal level dystrophin (Figure 5B). Western blot further confirmed low-level dystrophin expression in mdx3cv mice (Figure 5B).15
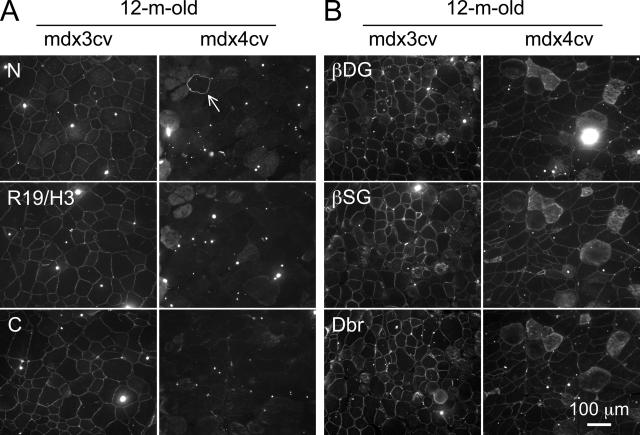
We observed a similar immunostaining profile in muscle samples obtained from 12-month-old mice. All myofibers in mdx3cv muscle expressed low-level dystrophin (Figure 6A). Except for rare revertant myofibers, dystrophin was not detected in mdx4cv muscle by the N-terminal, repeat 19/hinge 3, and C-terminal-specific antibodies. We also examined the representative components of the DGC by immunofluorescence staining. Similar to dystrophin immunostaining, we detected intense staining of β-dystroglycan, β-sarcoglycan, syntrophin, and dystrobrevin in BL6 muscle (Figures 5C and 6B). In mdx3cv muscle, we observed weak but continuous staining along the sarcolemma for all of the DGC components. Interestingly, we also found visible signals in mdx4cv muscle for all of the DGC components (Figures 5C and 6B).
Figure 6.
Characterization of dystrophin and DGC expression in 12-month-old mdx3cv and mdx4cv mice. A: Representative photomicrographs of immunofluorescence staining with epitope-specific antibodies that recognize N terminus (N), repeat 19/hinge 3 (R19/H3), and C terminus (C) of dystrophin. Arrow, a revertant myofiber in mdx4cv muscle. B: Representative photomicrographs of immunofluorescence staining with antibodies that recognize β-dystroglycan (βDG), β-sarcoglycan (βSG) and dystrobrevin (Dbr).
Utrophin Was Up-Regulated in Mdx3cv and Mdx4cv Muscles
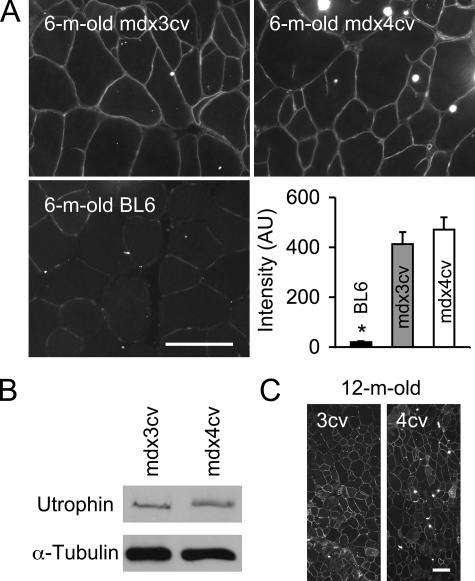
Utrophin up-regulation has been suggested as a compensatory mechanism in mdx muscle.25 A previous study also revealed utrophin up-regulation in mdx4cv mice.19 However, utrophin expression has not been evaluated in mdx3cv mice. To determine whether utrophin up-regulation contributed to the observed force preservation in mdx3cv mice, we compared utrophin expression in the EDL, TA, and gastrocnemius muscles as well as the diaphragm and the heart. As expected, we did not see sarcolemmal utrophin expression in BL6 muscle (TA muscle, Figure 7A). However, persistent utrophin up-regulation was found in both mdx3cv and mdx4cv muscles (Figure 7B and Supplementary Figure S1 at http://ajp.amjpathol.org). Furthermore, we did not see a significant difference between mdx3cv and mdx4cv muscles (Figure 7C).
Figure 7.
Up-regulation of utrophin in mdx3cv and mdx4cv skeletal muscles. A: Representative utrophin immunofluorescence staining in the TA muscles of 6-month-old mdx3cv, mdx4cv, and BL6 mice. Bar graph shows relative fluorescence signal intensity among three groups. AU, artificial unit. n = 6 for mdx3cv and BL6 muscles, n = 5 for mdx4cv muscle. Asterisk, value in BL6 muscle is significantly lower than that in mdx3cv and mdx4cv muscles. B: Representative utrophin Western blot from 6-month-old mdx3cv and mdx4cv skeletal muscle. C: Representative utrophin immunofluorescence staining in the TA muscles of 12-month-old mdx3cv and mdx4cv mice. Scale bars = 100 μm.
Discussion
DMD is a longstanding therapeutic challenge. Currently, there is no cure. Muscle damage and force reduction are two consequences of the loss of dystrophin. Experimental restoration of dystrophin has resulted in impressive muscle rescue in animal models of DMD. In these studies, improvement in muscle force is often accompanied by amelioration of muscle pathology. Furthermore, it is determined that an expression at one fifth of the wild-type dystrophin level is required to maintain normal muscle structure and function in mice.12,13 These results echo well with the findings in human patients. The relative level of dystrophin appears to be a key determinant of the clinical presentation.6,7,8,9,26 When there is equal or more than 20% of the normal dystrophin levels in muscle, patients usually show mild skeletal muscle disease. These patients are rarely confined to a wheelchair in their teenage years. A recent clinical study even suggests that 30% of the normal dystrophin level is enough to prevent skeletal muscle disease.27
Another group of patients has less than 20% dystrophin expression and they display intermediate phenotype.3,6,9,26 The mechanism(s) underlying the reduced clinical severity in these patients are not completely understood. Several hypotheses have been suggested including epigenetic, nutritional, and environmental factors, and a palliative effect of low-level dystrophin expression.9 A better understanding of the disease-modulating factors will help design novel DMD therapy.
Maintaining sarcolemma stability is a major function of dystrophin. From a mechanical standpoint, it is logical to speculate that a threshold dystrophin level is needed to provide the physical support and prevent the sarcolemma from tearing apart during contraction. This would predict an all-or-none effect. When the dystrophin level is less than the threshold level, muscle contraction rips the sarcolemma and initiates a chain reaction of muscle degeneration, inflammation, and eventually cell death. Based on this hypothesis, one would expect the less than threshold level dystrophin expression to be insignificant for therapy.
The classic mouse model for DMD is the mdx model. In this naturally occurring model, a nonsense mutation in exon 23 results in premature translation termination.28,29 The mutation also reduces dystrophin mRNA by nonsense mediated decay.30,31 Except for rare revertant myofibers, dystrophin expression is lost in mdx muscle. Chapman and colleagues14 generated several additional DMD mouse models using ENU mutagenesis. These models carry point mutations in different regions of the dystrophin gene. They either lead to premature translation termination (such as nonsense mutation in mdx4cv mice) or disrupt the conserved splicing sequences (such as in mdx2cv, mdx3cv, and mdx5cv mice).15,16 Among these, mdx4cv and mdx5cv are particularly attractive for stem cell and gene therapy studies because they have 10-fold fewer revertant fibers.17,18 Full-length dystrophin is not expressed in mdx4cv skeletal muscle.17,18,19 Interestingly, the point mutation in mdx3cv mice leads to multiple cryptic splicing events.15 The open reading frame is disrupted in the majority of the spliced products. However, a transcript that skips exons 65 and 66 translates into a near full-size dystrophin protein.15,17 Exons 65 and 66 encode the second half of the cysteine-rich domain. This domain anchors dystrophin to the sarcolemma through its interaction with β-dystroglycan. Surprisingly, the exon 65/66 deleted dystrophin protein still localizes to the sarcolemma.15,17,32 To further study the functional role of this internally truncated protein, Rafael and colleagues32 expressed the exon 65/66 deleted dystrophin in transgenic mdx mice. Interestingly, the membrane-bound mutant protein does not recruit β-dystroglycan and the sarcoglycans to the sarcolemma. Furthermore, it does not ameliorate the histological abnormalities.32
Several studies have shown dystrophic muscle pathology in mdx3cv and mdx4cv mice.15,17,18,19 Consistent with these reports, we also observed muscle hypertrophy (Table 1), variable myofiber size, central nucleation, macrophage infiltration, and sarcolemma damage in both mdx3cv and mdx4cv muscles (Figures 3 and 4). Collectively, these results suggest that a dystrophin level higher than what is presented in mdx3cv muscle is required to halt muscle degeneration.
Li and colleagues18 characterized muscle force in mdx4cv mice. These mice displayed similar force deficit as mdx mice. To elucidate the physiological consequence of the low-level near full-length dystrophin expression in mdx3cv mice, we measured the muscle force and the eccentric contraction-induced force decline in the EDL muscle (Figures 1 and 2). We also examined forelimb grip strength (Figure 3). In contrast to the lack of histopathology protection, low-level expression of this internally deleted dystrophin protein significantly improved specific tetanic force in both age groups when muscles were stimulated at 120 Hz and 150 Hz. In 6-month-old mice, the mdx3cv EDL muscle generated 23% more force per cross-sectional area than that of the mdx4cv EDL muscle (Figure 1B). In 12-month-old mice, the difference is reduced but the specific force of the mdx3cv EDL muscle was still 16% higher than that of the mdx4cv EDL muscle (Figure 2B). Consistent with the ex vivo assay with a single muscle, mdx3cv mice performed better in a forelimb grip strength assay. Mdx3cv mice generated 20 to 30% more force than that of age- and sex-matched mdx4cv mice. We also challenged the EDL muscle with eccentric contraction stress, the mdx3cv EDL muscle showed significantly better force preservation at 6 months of age (Figure 1C). A similar trend was observed in 12-month-old mice but it is not statistically significant (Figure 2C).
Rafael and colleagues33 previously compared wire maneuver performance in 2-month-old mdx and mdx3cv mice. They showed a delayed onset of muscle impairment in mdx3cv mice.33 The physiology findings described here provide direct evidence that muscle function is moderately preserved in mdx3cv mice. However, the limited protection seems to wear-out when mice get old. This profile seems to mimic clinical findings in BMD patients. We speculate that low-level (<20%) expression of a membrane-bound dystrophin may offer some force support. Consequently it may postpone wheelchair need in BMD patients. However, because muscle degeneration is not halted, patients eventually loss their muscle and die from muscular dystrophy.
It is currently not clear why a dystrophin that fails to restore the DGC on its own improves muscle force. One possibility is a synergistic effect between the up-regulated utrophin and low-level dystrophin expression. As shown in mdx4cv muscle, utrophin up-regulation alone is apparently insufficient to improve muscle force. The combined effect of utrophin up-regulation and low-level membrane-bound dystrophin may have tipped the balance in favor of force preservation. Future studies in the utrophin-null background are needed to clarify this issue. Such studies may also offer clues to whether low-level dystrophin expression affects other aspects of the disease, in particular, the lifespan. Another possibility is that the internally deleted dystrophin may interact with other yet undefined membrane protein(s) and/or cytoskeleton protein(s). These interactions may lead to muscle force preservation in mdx3cv mice.
Acknowledgments
We thank Ms. Chun Long for excellent technical assistance, Mr. Brian Bostick for proofreading and editing, and Drs. Glenn Morris and Lam Le for providing dystrophin monoclonal antibodies.
Footnotes
Address reprint requests to Dongsheng Duan, Ph.D., Associate Professor, Department of Molecular Microbiology and Immunology, The University of Missouri, School of Medicine, One Hospital Dr. M610G, MSB, Columbia, MO 65212. E-mail: duand@missouri.edu.
Supported by the National Institutes of Health (grant AR-49419 to D.D.) and the Muscular Dystrophy Association (to D.D.).
Supplementary material for this article can be found on http://ajp.amjpathol.org.
References
- Kunkel LM. 2004 William Allan Award Address. Cloning of the DMD gene. Am J Hum Genet. 2005;76:205–214. doi: 10.1086/428143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emery AEH, Muntoni F. Emery AEH, Muntoni F, editors. New York: Oxford University Press,; Duchenne Muscular Dystrophy. 2003:pp 26–44. [Google Scholar]
- Hoffman EP, Fischbeck KH, Brown RH, Johnson M, Medori R, Loike JD, Harris JB, Waterston R, Brooke M, Specht L, Kupsky W, Chamberlain J, Caskey Y, Shapiro F, Kunkel LM. Characterization of dystrophin in muscle-biopsy specimens from patients with Duchenne’s or Becker’s muscular dystrophy. N Engl J Med. 1988;318:1363–1368. doi: 10.1056/NEJM198805263182104. [DOI] [PubMed] [Google Scholar]
- England SB, Nicholson LV, Johnson MA, Forrest SM, Love DR, Zubrzycka-Gaarn EE, Bulman DE, Harris JB, Davies KE. Very mild muscular dystrophy associated with the deletion of 46% of dystrophin. Nature. 1990;343:180–182. doi: 10.1038/343180a0. [DOI] [PubMed] [Google Scholar]
- Harper SQ, Hauser MA, DelloRusso C, Duan D, Crawford RW, Phelps SF, Harper HA, Robinson AS, Engelhardt JF, Brooks SV, Chamberlain JS. Modular flexibility of dystrophin: implications for gene therapy of Duchenne muscular dystrophy. Nat Med. 2002;8:253–261. doi: 10.1038/nm0302-253. [DOI] [PubMed] [Google Scholar]
- Hoffman EP, Kunkel LM, Angelini C, Clarke A, Johnson M, Harris JB. Improved diagnosis of Becker muscular dystrophy by dystrophin testing. Neurology. 1989;39:1011–1017. doi: 10.1212/wnl.39.8.1011. [DOI] [PubMed] [Google Scholar]
- Bulman DE, Murphy EG, Zubrzycka-Gaarn EE, Worton RG, Ray PN. Differentiation of Duchenne and Becker muscular dystrophy phenotypes with amino- and carboxy-terminal antisera specific for dystrophin. Am J Hum Genet. 1991;48:295–304. [PMC free article] [PubMed] [Google Scholar]
- Byers TJ, Neumann PE, Beggs AH, Kunkel LM. ELISA quantitation of dystrophin for the diagnosis of Duchenne and Becker muscular dystrophies. Neurology. 1992;42:570–576. [PubMed] [Google Scholar]
- Beggs AH, Hoffman EP, Snyder JR, Arahata K, Specht L, Shapiro F, Angelini C, Sugita H, Kunkel LM. Exploring the molecular basis for variability among patients with Becker muscular dystrophy: dystrophin gene and protein studies. Am J Hum Genet. 1991;49:54–67. [PMC free article] [PubMed] [Google Scholar]
- Hoffman EP. Genotype/phenotype correlations in Duchenne/Becker dystrophy. Mol Cell Biol Hum Dis Ser. 1993;3:12–36. doi: 10.1007/978-94-011-1528-5_2. [DOI] [PubMed] [Google Scholar]
- Wells DJ, Wells KE, Walsh FS, Davies KE, Goldspink G, Love DR, Chan-Thomas P, Dunckley MG, Piper T, Dickson G. Human dystrophin expression corrects the myopathic phenotype in transgenic mdx mice. Hum Mol Genet. 1992;1:35–40. doi: 10.1093/hmg/1.1.35. [DOI] [PubMed] [Google Scholar]
- Wells DJ, Wells KE, Asante EA, Turner G, Sunada Y, Campbell KP, Walsh FS, Dickson G. Expression of human full-length and minidystrophin in transgenic mdx mice: implications for gene therapy of Duchenne muscular dystrophy. Hum Mol Genet. 1995;4:1245–1250. doi: 10.1093/hmg/4.8.1245. [DOI] [PubMed] [Google Scholar]
- Phelps SF, Hauser MA, Cole NM, Rafael JA, Hinkle RT, Faulkner JA, Chamberlain JS. Expression of full-length and truncated dystrophin mini-genes in transgenic mdx mice. Hum Mol Genet. 1995;4:1251–1258. doi: 10.1093/hmg/4.8.1251. [DOI] [PubMed] [Google Scholar]
- Chapman VM, Miller DR, Armstrong D, Caskey CT. Recovery of induced mutations for X chromosome-linked muscular dystrophy in mice. Proc Natl Acad Sci USA. 1989;86:1292–1296. doi: 10.1073/pnas.86.4.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox GA, Phelps SF, Chapman VM, Chamberlain JS. New mdx mutation disrupts expression of muscle and nonmuscle isoforms of dystrophin. Nat Genet. 1993;4:87–93. doi: 10.1038/ng0593-87. [DOI] [PubMed] [Google Scholar]
- Im WB, Phelps SF, Copen EH, Adams EG, Slightom JL, Chamberlain JS. Differential expression of dystrophin isoforms in strains of mdx mice with different mutations. Hum Mol Genet. 1996;5:1149–1153. doi: 10.1093/hmg/5.8.1149. [DOI] [PubMed] [Google Scholar]
- Danko I, Chapman V, Wolff JA. The frequency of revertants in mdx mouse genetic models for Duchenne muscular dystrophy. Pediatr Res. 1992;32:128–131. doi: 10.1203/00006450-199207000-00025. [DOI] [PubMed] [Google Scholar]
- Li S, Kimura E, Ng R, Fall BM, Meuse L, Reyes M, Faulkner JA, Chamberlain JS. A highly functional mini-dystrophin/GFP fusion gene for cell and gene therapy studies of Duchenne muscular dystrophy. Hum Mol Genet. 2006;15:1610–1622. doi: 10.1093/hmg/ddl082. [DOI] [PubMed] [Google Scholar]
- Judge LM, Haraguchiln M, Chamberlain JS. Dissecting the signaling and mechanical functions of the dystrophin-glycoprotein complex. J Cell Sci. 2006;119:1537–1546. doi: 10.1242/jcs.02857. [DOI] [PubMed] [Google Scholar]
- Lai Y, Yue Y, Liu M, Ghosh A, Engelhardt JF, Chamberlain JS, Duan D. Efficient in vivo gene expression by trans-splicing adeno-associated viral vectors. Nat Biotechnol. 2005;23:1435–1439. doi: 10.1038/nbt1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Y, Liu M, Duan D. C-terminal truncated microdystrophin recruits dystrobrevin and syntrophin to the dystrophin-associated glycoprotein complex and reduces muscular dystrophy in symptomatic utrophin/dystrophin double knock-out mice. Mol Ther. 2006;14:79–87. doi: 10.1016/j.ymthe.2006.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanh LT, Nguyen TM, Helliwell TR, Morris GE. Characterization of revertant muscle fibers in Duchenne muscular dystrophy, using exon-specific monoclonal antibodies against dystrophin. Am J Hum Genet. 1995;56:725–731. [PMC free article] [PubMed] [Google Scholar]
- Yue Y, Li Z, Harper SQ, Davisson RL, Chamberlain JS, Duan D. Microdystrophin gene therapy of cardiomyopathy restores dystrophin-glycoprotein complex and improves sarcolemma integrity in the mdx mouse heart. Circulation. 2003;108:1626–1632. doi: 10.1161/01.CIR.0000089371.11664.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Yue Y, Harper SQ, Grange RW, Chamberlain JS, Duan D. Adeno-associated virus-mediated micro-dystrophin expression protects young Mdx muscle from contraction-induced injury. Mol Ther. 2005;11:245–256. doi: 10.1016/j.ymthe.2004.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst RC, McCullagh KJ, Davies KE. Utrophin upregulation in Duchenne muscular dystrophy. Acta Myol. 2005;24:209–216. [PubMed] [Google Scholar]
- Tuffery-Giraud S, Saquet C, Thorel D, Disset A, Rivier F, Malcolm S, Claustres M. Mutation spectrum leading to an attenuated phenotype in dystrophinopathies. Eur J Hum Genet. 2005;13:1254–1260. doi: 10.1038/sj.ejhg.5201478. [DOI] [PubMed] [Google Scholar]
- Neri M, Torelli S, Brown S, Ugo I, Sabatelli P, Merlini L, Spitali P, Rimessi P, Gualandi F, Sewry C, Ferlini A, Muntoni F. Dystrophin levels as low as 30% are sufficient to avoid muscular dystrophy in the human. Neuromuscul Disord. 2007;17:913–918. doi: 10.1016/j.nmd.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Bulfield G, Siller WG, Wight PA, Moore KJ. X chromosome-linked muscular dystrophy (mdx) in the mouse. Proc Natl Acad Sci USA. 1984;81:1189–1192. doi: 10.1073/pnas.81.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicinski P, Geng Y, Ryder-Cook AS, Barnard EA, Darlison MG, Barnard PJ. The molecular basis of muscular dystrophy in the mdx mouse: a point mutation. Science. 1989;244:1578–1580. doi: 10.1126/science.2662404. [DOI] [PubMed] [Google Scholar]
- Chamberlain JS, Pearlman JA, Muzny DM, Gibbs RA, Ranier JE, Caskey CT, Reeves AA. Expression of the murine Duchenne muscular dystrophy gene in muscle and brain. Science. 1988;239:1416–1418. doi: 10.1126/science.3347839. [DOI] [PubMed] [Google Scholar]
- Buvoli M, Buvoli A, Leinwand LA. Interplay between exonic splicing enhancers, mRNA processing, and mRNA surveillance in the dystrophic Mdx mouse. PLoS ONE. 2007;2:e427. doi: 10.1371/journal.pone.0000427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafael JA, Cox GA, Corrado K, Jung D, Campbell KP, Chamberlain JS. Forced expression of dystrophin deletion constructs reveals structure-function correlations. J Cell Biol. 1996;134:93–102. doi: 10.1083/jcb.134.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafael JA, Nitta Y, Peters J, Davies KE. Testing of SHIRPA, a mouse phenotypic assessment protocol, on Dmd(mdx) and Dmd(mdx3cv) dystrophin-deficient mice. Mamm Genome. 2000;11:725–728. doi: 10.1007/s003350010149. [DOI] [PubMed] [Google Scholar]