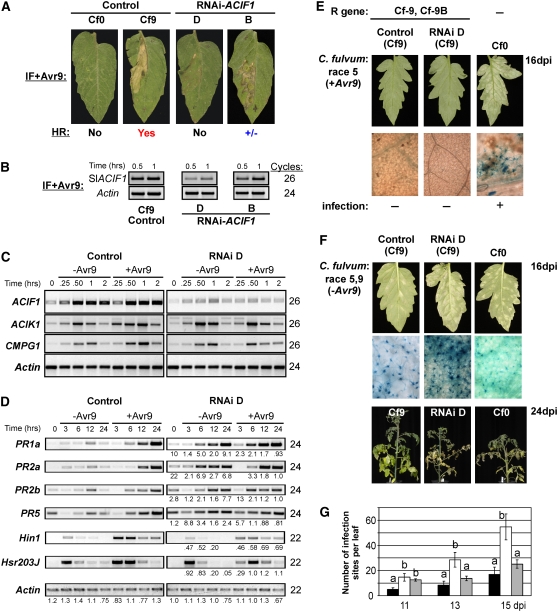
Figure 8.
RNAi-Mediated Silencing of Tomato ACIF1 Leads to the Loss of Cf-9B, but Not Cf-9, Resistance to C. fulvum.
(A) HR at 2 d after infiltration of IF+Avr9 in leaves of tomato RNAi-ACIF1 (lines B and D) compared with the negative and positive control plants (MM Cf0 and MM Cf9 tomato, respectively).
(B) RT-PCR analysis of ACIF1 transcript levels in tomato RNAi-ACIF1 (lines B and D) and a control after IF+Avr9 infiltration (0.5 and 1 h after infiltration). Tomato actin transcript levels are shown as a control for equal cDNA levels. The number of cycles used during PCR amplification is shown.
(C) RT-PCR analysis of leaf material at the indicated time points after infiltration with IF+Avr9 or IF−Avr9 (mock). Transcript levels were determined for ACIF1, ACIK1, and CMPG1. Actin transcript levels are shown as a control for equal cDNA levels. The number of PCR cycles is indicated at right for each primer combination.
(D) RT-PCR of PR genes (PR1A, PR2A, PR2B, and PR5) and HR marker genes (Hin1 and Hsr203J) in tomato RNAi-ACIF1 (line D) and the control after infiltration with IF−Avr9 or IF+Avr9. The relative difference in expression levels between the control and the RNAi D line are indicated below the panel for the different time points (the number signifies the fold change in signal intensity).
(E) Typical leaves (top panels) of 4-week-old tomato plants (cv MM; susceptible Cf0, resistant Cf9, and Cf9 transgenic for RNAi-ACIF1 D) at 16 DAI with C. fulvum race 5 (+Avr9). Conidiophores (blue threads, bottom panel) emerged through the stomata only in the case of a successful infection (Cf0), as microscopically confirmed after trypan blue staining.
(F) The same as in (E) except that plants were infected with C. fulvum race 5.9 (−Avr9). The plants are also shown at 24 DPI. This experiment was repeated four times with similar results.
(G) Mean (±se) number of infection sites macroscopically observed at 11, 13, and 15 DPI after infection with race 5.9 on Cf0 (gray bars), Cf9 (black bars), or RNAi-ACIF1 (white bars). Six plants were infected per line, and we counted the number of infection patches on two leaves for all plants per time point. Means were compared by Student's t test. Different letters above two columns indicate significantly different means at a single time point (P < 0.05). This experiment was independently repeated four times with similar results.