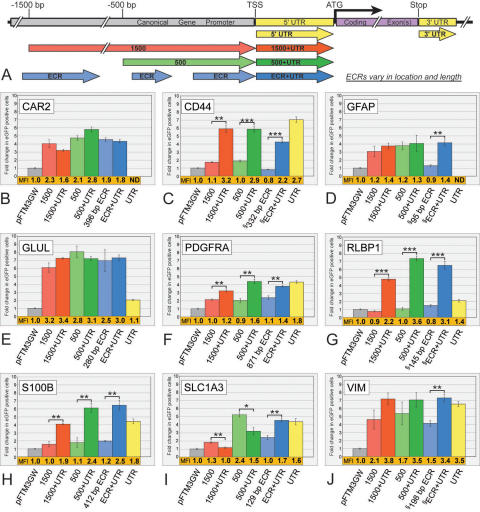
Figure 2.
Flow cytometry analysis of promoter fragments from nine Müller cell expressed genes. A: Diagrammatic representation of DNA fragments analyzed here and in subsequent figures. B-J: Significant variability in both number of positive cells and fluorescence intensity are apparent among the 61 fragments tested. Seven fragments were analyzed for each gene: the 5′ UTR alone (yellow), a ~1500 bp (red), a ~500 bp (green), and a variable-sized evolutionarily conserved region (ECR; blue), each with and without their respective 5′ UTRs. Bars indicate fold change in number of eGFP positive cells, normalized to the promoter-less parent vector (pFTM3GW). The mean fluorescence intensity (MFI) for each construct was also normalized to the parent vector (pFTM3GW) and is shown immediately below each bar (shown in orange). UTRs for the GFAP (14 bp) and CAR2 (28 bp) genes were quite short and not individually tested. Both the locationrelative to the transcriptional start site (TSS), and the size of the ECRs vary for each gene (official gene names can be found in Introduction). Refer to Appendix 1 for genomic coordinates of each construct. The symbol (§) identifies ECRs that were not immediately adjacent to the TSS (CD44, GFAP, RLBP1, and VIM), and less-conserved DNA between the ECR and TSS was included for each of these constructs. ATG is the codon for the starting methionine. Error bars represent 1 standard deviation. The single asterisk equals p<0.01, the double asterisk equals p<0.001, and the triple asterisk equals p<0.0001, using a two-tailed Student's t-test assuming equal variances. ND means “not determined.”