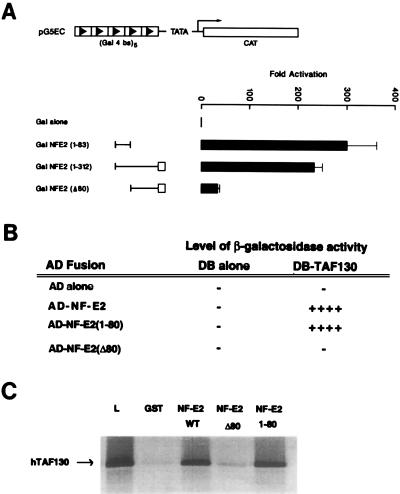
Figure 3.
Colocalization of the activation domain of NF-E2 with the TAFII130 binding domain. (A) Localization of activation domain of NF-E2. K562 cells were cotransfected by electroporation with expression vectors for GAL4 fusion proteins containing the indicated regions of NF-E2 and a reporter construct containing five GAL4 DNA binding sites regulating the expression of the chloramphenicol acetyltransferase gene (pG5EC). The first 312 amino acids of NF-E2 previously have been shown to contain a proline-rich activation domain lacking structural homology to other proteins denoted by a line, and a basic region similar to other cap’n-collar transcriptional activators denoted by an open box. The expression vector used in this study was pCINeo, containing the cytomegalovirus promoter. The fold activation represents the chloramphenicol acetyltransferase activity relative to the expression of the expression vector containing the GAL4 DNA binding domain alone. (B) hTAFII130 interacts with the NF-E2 activation domain in the yeast two-hybrid assay. The S. cerevisiae reporter strain SFY526, which contains a β-galactosidase reporter construct under the control of a GAL4 binding motif multimer, was transformed with the indicated plasmids. The AD-NF-E2 constructs contain the stated regions of the coding sequence of human NF-E2 inserted in-frame to the activation domain of GAL4 (amino acids 768–881). DB-TAFII130 contains the partial coding sequence of the human protein inserted in-frame to the GAL4 DNA binding domain (amino acids 1–147). (C) The amino terminal 80 amino acids of NF-E2 interacts specifically with hTAFII130 in the GST chromatography system. Fusion proteins containing GST fused to amino acids 1–374, 1–80, or 80–374 of NF-E2 or GST alone were expressed in Escherichia coli and bound to glutathione-Sepharose beads. The beads then were incubated with 35S-labeled in vitro-translated hTAFII130. Other experimental details are as stated in Fig. 2C.