To elucidate the mechanism of regulation of aspartate kinase, the regulatory subunit (the β subunit of T. thermophilus aspartate kinase) was crystallized in the presence of the inhibitor threonine.
Keywords: aspartate kinase, amino-acid biosynthesis
Abstract
Aspartate kinase (AK) from Thermus thermophilus, which catalyzes the first step of threonine and methionine biosynthesis, is regulated via feedback inhibition by the end product threonine. To elucidate the mechanism of regulation of AK, the regulatory subunit (the β subunit of T. thermophilus AK) was crystallized in the presence of the inhibitor threonine. Diffraction data were collected to 2.15 Å at a synchrotron source. The crystal belongs to the cubic space group P4332 or P4132, with unit-cell parameters a = b = c = 141.8 Å.
1. Introduction
Aspartate kinase (AK) catalyzes the phosphorylation of aspartic acid, the first step in the biosynthesis of the aspartic amino-acid family lysine, threonine and methionine. Similar to other enzymes involved in the first steps of amino-acid biosynthesis, AK is regulated through feedback inhibition by the end products. AK from Thermus thermophilus AT-62 (formerly named T. flavus AT-62) is inhibited by threonine (Nishiyama et al., 1995 ▶), while AK from Corynebacterium glutamicum is inhibited by lysine and threonine in a concerted manner (Shiio & Miyajima, 1969 ▶). AK is also important for industrial production of the amino acids belonging to the aspartic amino-acid family (Tosaka et al., 1983 ▶; Jetten & Sinskey, 1995 ▶), which are essential for mammals. In addition to these essential amino acids, diaminopimelic acid, an intermediate in lysine biosynthesis, is a key compound that is necessary for cell-wall synthesis in most bacteria. Therefore, an enzyme that is involved in diaminopimelic acid biosynthesis may serve as a target for the design of antimicrobial agents, especially for pathogenic bacteria (Patte, 1996 ▶; Girodeau et al., 1986 ▶). Thus, AK is an attractive target for both scientific understanding and industrial applications. AK from T. thermophilus is composed of two subunits, α and β, which are encoded by an in-frame overlapping gene (Nishiyama et al., 1995 ▶) as in C. glutamicum AK and Bacillus subtilis AK II to form an α2β2 tetramer (Kalinowski et al., 1991 ▶; Chen et al., 1987 ▶). The β subunit is identical to about 160 amino acids of the C-terminus of the α subunit. In this α2β2-type AK, the N-terminal regions of the α subunit serve as catalytic domains and the C-terminal region of the α subunit and the β subunit function as regulatory domains (Kobashi et al., 1999 ▶; Marco-Marín et al., 2003 ▶; Kato et al., 2004 ▶). Aravind and Koonin discovered a motif that is conserved in many allosteric enzymes involved in amino-acid and purine biosynthesis and named the motif the ‘ACT domain’ as an acronym for aspartate kinase, chorismate mutase and TyrA (prephenate dehydrogenase; Aravind & Koonin, 1999 ▶). The motif was expected to serve as a small-molecule-binding domain for catalytic regulation, forming a β-α-β-β-α-β fold (Schuller et al., 1995 ▶). The crystal structures of several homo-oligomeric AKs have recently been reported (Mas-Droux et al., 2006 ▶; Kotaka et al., 2006 ▶; Faehnle et al., 2006 ▶). In contrast to these AKs, which have dimeric quaternary structures in which two ACT domains from each subunit interact with each other to form a dimer, AK from T. thermophilus has a different α2β2 subunit organization in which a single β subunit of α2β2-type AK contains two ACT-domain motifs. The amino-acid sequence identity of the β subunit of T. thermophilus AK is 20%, 17% and 27% to the ACT domain-containing portions of aspartate kinase I from Arabidopsis thaliana, aspartate kinase III from Escherichia coli and aspartate kinase from Methanococcus jannashii, respectively. This suggests that it has a different mechanism for subunit assembly and regulation. To elucidate the regulatory mechanism, we crystallized the regulatory subunit (the β subunit) of T. thermophilus AK.
2. Experimental
2.1. Protein expression and purification
The askB gene (accession No. D37928) encoding the β subunit of T. thermophilus AK was amplified by polymerase chain reaction using the oligonucleotides 3′-GGGGAATTCTCAAGGAGGTGTCATATGGAGATGGACAAGGCG-5′ and 3′-CCCAAGCTTTCAGTGGTGGTGGTGGTGGTGGGCCTTGTCCAGCTC-5′. The amplified DNA fragment designed to direct the production of the full-length β subunit (Met1–Ala161) with a His6-tag extension at the C-terminal end was cloned into the EcoRI/HindIII site of pBluescriptII SK(+). The calculated theoretical molecular weight of the protein is 17 717 Da. After verifying the nucleotide sequence, the DNA fragment was cloned into the NdeI/HindIII site of pET26b(+) and introduced into E. coli BL21-CodonPlus(DE3)-RIL cells. The cells were grown in 2×YT broth in the presence of kanamycin (50 µg ml−1) and chloramphenicol (30 µg ml−1) at 303 K. When the optical density at 600 nm of the culture reached about 0.6, gene expression was induced by adding 0.1 mM isopropyl β-d-thiogalactopyranoside and the culture was continued for an additional 12–14 h. The cells were harvested and washed in buffer A (20 mM Tris–HCl pH 7.5) and suspended in buffer B (20 mM Tris–HCl pH 7.5, 150 mM NaCl). Suspended cells were disrupted by sonication and centrifuged at 40 000g for 25 min. The supernatant was applied onto an Ni-resin column (Clontech) equilibrated with buffer B supplemented with 20 mM imidazole. After washing with buffer B containing 20 mM imidazole and successive washing with buffer B containing 50 mM imidazole, the proteins bound to the resin were eluted with buffer B containing 200 mM imidazole and then with buffer B containing 500 mM imidazole. The flow rate and fraction volume for the nickel-affinity chromatography were 1 ml min−1 and 1 ml, respectively. Fractions containing the His6-tagged β subunit were mixed and concentrated to about 40 mg ml−1 using Vivaspin-20 centrifugal filtration with a 10 kDa cutoff (Vivascience). The concentrated sample was applied onto a HiLoad 26/60 Superdex 75 gel-filtration FPLC column (Amersham Bioscience) equilibrated with buffer B and fractions were collected every 2 min using a flow rate of 2.5 ml min−1. Based on the elution volume from the size-exclusion column, the oligomeric state of the protein was estimated to be a monomer. The >95% homogeneity of the purified β subunit was verified by SDS–PAGE. Over 60 mg of the β subunit of T. thermophilus AK with a His6 tag was purified from 1 l culture and was used for crystallization without removal of the tag.
2.2. Crystallization
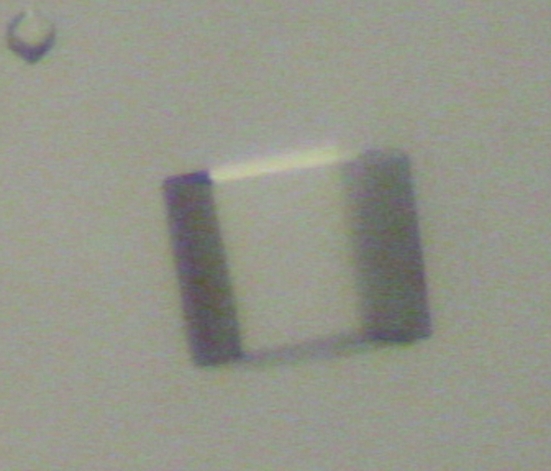
Crystallization conditions were screened by the hanging-drop vapour-diffusion method using Crystal Screen kits (Hampton Research). 2 µl drops consisting of 1 µl reservoir solution and 1 µl 10 mg ml−1 β-subunit solution with or without 5 mM threonine were equilibrated against 500 µl reservoir solution at 293 K. A few crystals were obtained from threonine-containing droplets using solution No. 9 (0.1 M sodium acetate trihydrate pH 4.6, 2.0 M sodium chloride) from Crystal Screen II. Crystals with dimensions of 0.30 × 0.30 × 0.30 mm formed in 0.1 M sodium acetate trihydrate pH 4.6 and 1.2–2.0 M sodium chloride (Fig. 1 ▶) were used for X-ray diffraction.
Figure 1.
Cubic crystal of the β subunit of T. thermophilus AK.
2.3. Data collection and processing
Prior to data collection, crystals were briefly soaked in a cryoprotectant solution containing the same concentrations of sodium acetate and sodium chloride as in the crystallization condition and 25%(v/v) glycerol, flash-cooled in a nitrogen-gas stream at 95 K and stored in liquid nitrogen. Diffraction data (λ = 1.000 Å) were collected using a charge-coupled device (CCD) camera (ADSC Quantum 210) with a crystal-to-detector distance of 189.60 mm, an oscillation angle of 0.5°, a total of 180 images and an exposure time of 2.5 s at the NW12A station of the Photon Factory AR, High Energy Accelerator Research Organization (KEK), Tsukuba, Japan. Diffraction images were indexed, integrated and scaled using the HKL-2000 program suite (Otwinowski & Minor, 1997 ▶). The crystal belongs to the cubic space group P4332 or P4332, with unit-cell parameters a = b = c = 141.8 Å. Assuming the presence of two monomers of 18 kDa protein in the asymmetric unit, the calculated Matthews coefficient (V M) is 3.3 Å3 Da−1, with a solvent content of 63.0%. A complete data set has been obtained to 2.15 Å, corresponding to an R merge of 6.7%. Details of the data-collection statistics are summarized in Table 1 ▶. We are now in the process of attempting to use the multiwavelength anomalous diffraction method to solve the three-dimensional structure of the β subunit of T. thermophilus AK. Solving this structure will provide the first structure of the regulatory subunit of an α2β2-type aspartate kinase.
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| Beamline | NW12A |
| Space group | P4132 or P4332 |
| Unit-cell parameters (Å) | a = b = c = 141.8 |
| Resolution (Å) | 2.15 (2.15–2.19) |
| Total reflections | 578149 |
| Unique reflections | 27145 |
| Rmerge† (%) | 6.7 |
| I/σ(I) | 59.9 (11.1) |
| Completeness | 100.0 (100.0) |
R
merge = 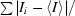
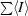 .
.
Acknowledgments
We would like to thank Dr Elinor T. Adman (University of Washington School of Medicine) for her assistance in completing the manuscript. We also appreciate the staff of the Photon Factory for their assistance with the data collection. This work was performed under the approval of the Photon Factory Program Advisory Committee (Proposal No. 05 G268).
References
- Aravind, L. & Koonin, E. V. (1999). J. Mol. Biol.287, 1023–1040. [DOI] [PubMed] [Google Scholar]
- Chen, N. Y., Hu, F. M. & Paulus, H. (1987). J. Biol. Chem.262, 8787–8798. [PubMed] [Google Scholar]
- Faehnle, C. R., Liu, X., Pavlovsky, A. & Viola, R. E. (2006). Acta Cryst. F62, 962–966. [DOI] [PMC free article] [PubMed]
- Girodeau, J. M., Agouridas, C., Masson, M., Pineau, R. & Le Goffic, F. (1986). J. Med. Chem.29, 1023–1030. [DOI] [PubMed] [Google Scholar]
- Jetten, M. S. & Sinskey, A. J. (1995). Crit. Rev. Biotechnol.15, 73–103. [DOI] [PubMed] [Google Scholar]
- Kalinowski, J., Cremer, J., Bachmann, B., Eggeling, L., Sahm, H. & Pühler, A. (1991). Mol. Microbiol.5, 1197–1204. [DOI] [PubMed] [Google Scholar]
- Kato, C., Kurihara, T., Kobashi, N., Yamane, H. & Nishiyama, M. (2004). Biochem. Biophys. Res. Commun.316, 802–808. [DOI] [PubMed] [Google Scholar]
- Kobashi, N., Nishiyama, M. & Tanokura, M. (1999). J. Biosci. Bioeng.87, 739–745. [DOI] [PubMed] [Google Scholar]
- Kotaka, M., Ren, J., Lockyer, M., Hawkins, A. R. & Stammers, D. K. (2006). J. Biol. Chem.281, 31544–31552. [DOI] [PubMed] [Google Scholar]
- Marco-Marín, C., Ramón-Maiques, S., Tavárez, S. & Rubio, V. (2003). J. Mol. Biol.334, 459–476. [DOI] [PubMed] [Google Scholar]
- Mas-Droux, C., Curien, G., Robert-Genthon, M., Laurencin, M., Ferrer, J. L. & Dumas, R. (2006). Plant Cell, 18, 1681–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama, M., Kukimoto, M., Beppu, T. & Horinouchi, S. (1995). Microbiology, 141, 1211–1219. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Patte, J. (1996). Escherichia Coli and Salmonella: Cellular and Molecular Biology, 2nd ed., edited by F. C. Neidhardt, pp. 528–541. Washington DC: American Society for Microbiology.
- Schuller, D. J., Grant, G. A. & Banaszak, L. J. (1995). Nature Struct. Biol.2, 69–76. [DOI] [PubMed] [Google Scholar]
- Shiio, I. & Miyajima, R. (1969). J. Biochem. (Tokyo), 65, 849–859. [DOI] [PubMed] [Google Scholar]
- Tosaka, O., Enei, H. & Hirose, Y. (1983). Trends Biotechnol.1, 70–74.