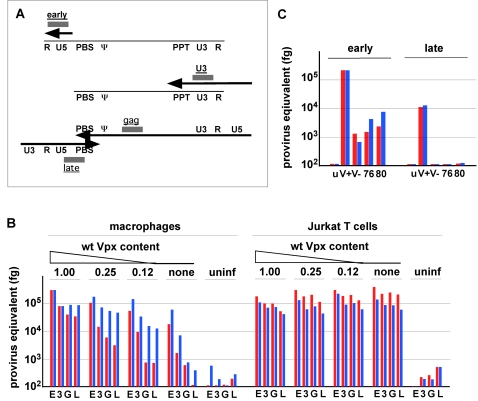
Figure 5. Reverse transcription of SIVmac 239(GFP) virions comprising Vpx(Q76A) and Vpx(F80A) in macrophages is compromised.
(A) Location of reverse transcription intermediates used to gauge the progression of reverse transcription of SIVmac 239(GFP) genome. Regions amplified by “early”, “U3, “gag” and “late” sets of oligonucleotide primers are represented by grey boxes. The thin line represents viral RNA and the locations of the R, U5, primer binding site (PBS), packaging signal (Ψ), polypurine tract (PPT) and U3 regions are indicated. The thick arrows represent viral cDNA. (B) Steady state levels of reverse transcription intermediates following infection with VSV-G pseudotyped single cycle SIVmac 239(GFP) virions deficient in Vpx. Macrophages and Jurkat T cells were infected with the reference set of SIVmac 239(GFP) virions containing various amounts of wild type Vpx, characterized in Figure 3. DNA was prepared from the infected cells 18 hour or 72 hours following infection and 50 ng aliquots were analyzed in duplicate by real time PCR with oligonucleotide primers that recognize “early”(E), “U3”(3), “gag”(G) and “late”(L) reverse transcription products, as indicated below the histograms. The amounts of reverse transcription products were calculated by comparison to standard curves generated with serially diluted SIVmac 239 proviral DNA and are shown in red and blue for the 18-hour and 72-hour timepoint, respectively. The variances between duplicate data points were less than 4%. (C) Q76A and F80A substitutions disrupt Vpx function in macrophages. Macrophages were not infected (u) or infected with single cycle SIVmac 239(GFP) virions containing Q76A (76) or F80A (80) substituted, wild type (V+), or no Vpx (V−), characterized in Figure 3A, and reverse transcription was analyzed with “early” and “late” primers as described above.