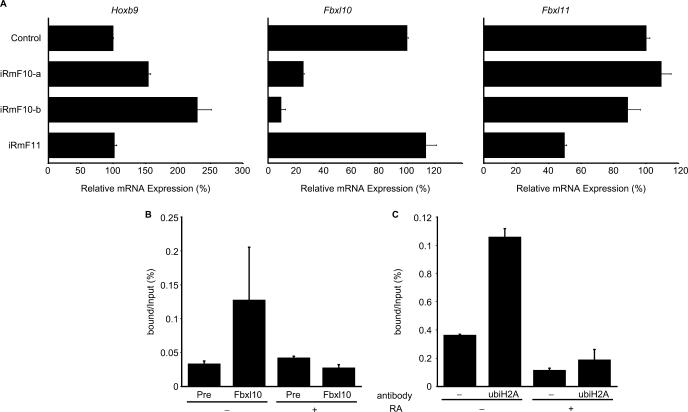
Figure 5.
Influence of Fbxl genes on the native Hoxb9 induction system. (A) The influences of Fbxl genes were analysed by introducing siRNAs. We designed three siRNAs—iRmF10-a, iRmF10-b and iRmF11—and used RT–PCR to examine the effects of these siRNAs on intrinsic Hoxb9 and Fbxl expression. The siRNAs were introduced into P19 cells during the 24-h pre-incubation period and during the 48-h RA-induction period (total exposure time: 72 h). RT–PCR samples were normalized relative to β-actin expression. Transfection of Fbxl10-specific siRNAs elevated Hoxb9 expression (left graph), whereas transfection of Fbxl11-specific siRNAs did not significantly influence Hoxb9 expression. The specificity of these siRNAs on target genes is shown in the remaining graphs as indicated. (B) Binding of FBXL10 protein to native Hoxb9 promoter in P19 cells. ChIP analysis revealed that FBXL10 binds Hoxb9 promoter in undifferentiated P19 cells but not in differentiated P19 cells, which express Hoxb9. (C) Histone H2A ubiquitylation status of the Hoxb9 promoter differs as P19 cells differentiate. ChIP was carried out with anti-ubiquitylated histone H2A antibody. Similar to the profile observed for FBXL10, ubiquitylated histone H2A seems to accumulate at the Hoxb9 promoter region when P19 cells are in an undifferentiated state.