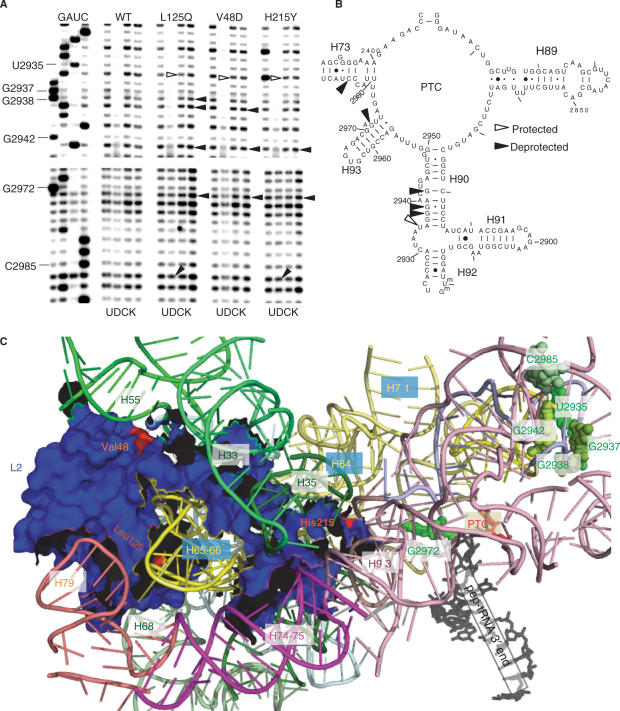
Figure 6.
Structure probing of wild-type and mutant ribosomes. (A) Autoradiograms of reverse transcriptase primer extension reactions spanning sequence in helices 90–73. Sequencing reactions (left sides of panels) are labeled corresponding to the rRNA sense strand. Below each panel, U stands for untreated, D is DMS, C denotes CMCT and K indicates kethoxal. Sources of ribosomes are indicated at top. Protected and deprotected bases are indicated by open and filled arrows, respectively. Chemical modification results in strong stops 1-nt 5′ of the base, and bases with altered chemical protection patterns are identified at left. (B) Localization of bases in the vicinity of the peptidyltransferase center (PTC) of yeast 25S rRNA whose chemical modification patterns were affected by the L2 mutants. (C) Mapping of the L2 mutants and rRNA protection data into the yeast ribosome structure generated by cryo-EM and fitted onto the H. marismortui large subunit crystal structure (13). L2, the PTC, the 3′ end of the peptidyl–tRNA and rRNA helices are labeled. The V48D and L125Q mutants map to the globular domain of L2 where they interact with helices 55 and 65/66, respectively. The H215Y mutant maps to the tip of the extended domain where it inserts into the major groove of helix 93. Nucleotides with altered chemical protection patterns are shown in shades of green. Base numbering follows the S. cerevisiae sequence shown in panel (B).