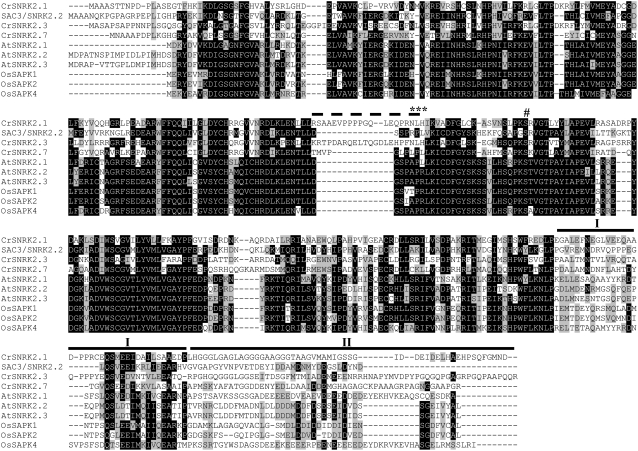
Figure 5.
Amino acid sequence alignment. The predicted SNRK2.1, SNRK2.2 (SAC3), SNRK2.3, and SNRK2.7 proteins were aligned with representative Arabidopsis and rice SNRK2 kinases (accession nos. as follows: AtSNRK2.1, NP_196476; AtSNRK2.2, NP_190619; AtSNRK2.3, NP_201489; SAPK1, NP_001050274; SAPK2, NP_001060312; SAPK4, NP_001044930) using BioEdit 7.0.5.3 software. The black and gray boxes indicate identical and similar amino acids, respectively. The C-terminal subdomains are highlighted with heavy black lines above the sequences and roman numerals above the line (Yoshida et al., 2006). The extra loop segment present in CrSNRK2.1 and CrSNRK2.3 (dotted line), the conserved NLH motif (***), and the conserved phosphorylated Ser of the activation domain (#) are noted.