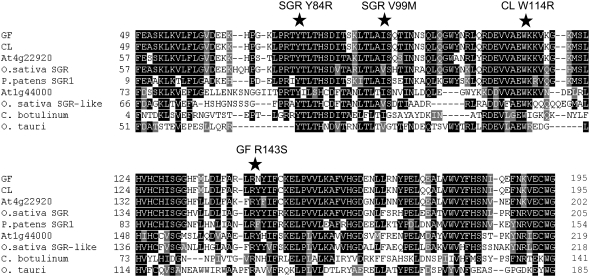
Figure 5.
Amino acid alignment of SGR homologs. Protein alignments based on the highly conserved central core of the SGR proteins were performed using ClustalW. Conserved amino acids are indicated by shaded squares. Highly conserved amino acids that, when mutated, give rise to mutant phenotypes (rice, Y84C and V99M; pepper, W114R; and tomato, R143S) are indicated by stars. Accession numbers are as follows (in parentheses): GF (EU414632); CL (EU414631); rice SGR (EAZ09856); P. patens SGR1 (EDQ70701); rice SGR-like (NP_001054370); Clostridium botulinum (YP_001391480); Oryza tauri (CAL56489). The Arabidopsis sequences At1g44000 and At4g22920 are based on TAIR annotations (http://www.arabidopsis.org).