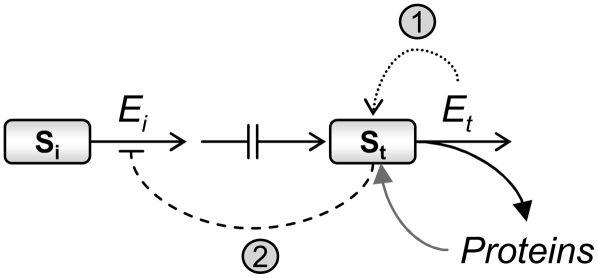
Figure 8.
Schematic representation of the proposed regulatory metabolic module. The regulatory steps are as follows. The major controller of the module is the gene encoding the catabolic enzyme Et, catalyzing the first catabolic step of the amino acid. Its upregulation stimulates the module by reducing the level of its substrate (St), namely, the amino acid (step 1; dotted curved arrow). Reduced levels of St stimulate the activity of allosteric biosynthetic Ei enzymes by reduction of its feedback inhibition by St (step 2; dashed curved line), stimulating the flux through the entire module. The broken line represents all biosynthetic/nonallosteric steps. The pool of the amino acid may also be determined by the extent of its incorporation into proteins (curved black arrow) and by the extent of protein breakdown (curved gray arrow). Our proposed module may also fit to actively growing (nonsenescence) tissues of plants grown under favorable (nonstress) conditions in which the catabolic enzymes are generally repressed, but the incorporation of the amino acids into proteins (black curved arrow) may transiently reduce the level of the amino acid (St) and as a consequence enhance the flux through the metabolic module by transiently reducing the feedback inhibition on the allosteric enzyme Ei.