Abstract
A cold-responsive chitinase gene, BiCHT1, was isolated from bromegrass (Bromus inermis) ‘Manchar’ suspension cells. BiCHT1 messenger RNA was detected at low levels in nonstressed bromegrass cells, whereas its accumulation was induced by incubation at 10°C and 4°C as detected by northern- and western-blot analyses. BiCHT1 was highly homologous to rye CHT9, known to encode an antifreeze protein. BiCHT1 was overexpressed in Escherichia coli and bromegrass cells using genetic transformation procedures. BiCHT1 products expressed in both systems had chitinase activity, but the expressed proteins did not affect the growth of ice crystals in any conditions tested. Besides cold stress, the expression of the BiCHT1 gene was up-regulated by exposure to 35°C, but not by salt or osmotic stress, abscisic acid, or ethephon. BiCHT1 messenger RNA did not accumulate in response to methyl jasmonate and salicylic acid, but was slightly increased by prolonged culture at 25°C and only transiently by chitin. Antifreeze activity detected in the culture medium was induced at 4°C but only slightly at 10°C. It was also induced by ethephon treatment, but not by abscisic acid, chitin, or prolonged incubation at 25°C. The results of transgenics and expression analyses suggest that the BiCHT1 product is a major protein with chitinase activity secreted in the medium of cold-treated cells and is unlikely to be responsible for the antifreeze activity detected in the culture medium.
Low temperature is a critical environmental factor that significantly influences plant growth, development, and crop yield. Most plants from temperate regions can acclimate to cold and develop freezing tolerance (Guy, 1990). Cold acclimation has been characterized by physiological and biochemical changes that result from the selective increases or decreases in the biosynthesis of a large number of distinct proteins (Bohnert et al., 1995). These changes in protein patterns are due in part to changes in the transcriptional activity of the corresponding genes. Many cold-inducible genes have been cloned from various plants (Thomashow, 1999). Some of these products are involved in the stabilization of membranes and macromolecules, metabolic shifts, scavenging of oxygen radicals, and signal transduction pathways. The function of numerous other cold-inducible products is still unknown. Analysis of genes whose expression is induced during cold acclimation is important to understand the mechanism of freezing tolerance and for use in breeding freezing-tolerant plants.
Many overwintering cereals accumulate antifreeze proteins (AFPs) during cold acclimation (Antikainen and Griffith, 1997). AFPs bind to the surface of ice crystals and retard and/or modify ice crystal growth (Ewart et al., 1999). In a similar fashion, AFPs inhibit ice recrystallization (Griffith and Yaish, 2004), which reduces ice crystal sublimation and maintains small ice crystals in frozen tissues. This prevents ice crystals from growing into tissue-damaging sizes during prolonged exposure to subzero temperatures and is thought to improve winter survival in the field (Chun et al., 1998). Based on these properties, AFPs are thought to play important roles in freezing tolerance and overwintering mechanisms. In cold-acclimated winter rye (Secale cereale) leaves, five major antifreeze proteins were present as the most abundant proteins in apoplastic extracts (Hon et al., 1994). Comparison of N-terminal amino acid sequences, immuno-cross reactions, and enzyme activity assays revealed that these antifreeze proteins are similar to pathogenesis-related proteins, such as chitinases, β-1,3-glucanases, and thaumatin-like proteins (Hon et al., 1995). Two different chitinase genes, named CHT9 and CHT46, were isolated from a cDNA library of cold-acclimated winter rye leaves (Yeh et al., 2000). CHT9 and CHT46 encoded class I chitinase and class II chitinase, respectively, and their products expressed in Escherichia coli modified the growth of ice crystals. However, to our knowledge, there has been no report on the overexpression of these products in plants or on the expression of these genes in plants under various conditions.
Chitinases (EC 3.2.1.14) hydrolyze chitin (N-acetyl-d-glucosamine polymers) and are present in many plant species. Plant chitinases are induced by pathogens or elicitors and are important enzymes in defense mechanisms. However, chitinases are also involved in diverse biological functions, such as somatic embryogenesis (De Jong et al., 1992), modification of ice crystal growth (Hon et al., 1995), and cell wall biogenesis (Mouille et al., 2003; D. Zhang et al., 2004).
In our previous study, BiCHT1 encoding a putative chitinase was isolated from bromegrass (Bromus inermis) ‘Manchar’ based on microarray analyses (T. Nakamura, J. Yazaki, N. Kishimoto, S. Kikuchi, and M. Ishikawa, unpublished data [hereafter referred to as Nakamura et al., unpublished data]). Expression of BiCHT1 mRNA was increased by low temperature, suggesting the product may be involved in cold acclimation. BiCHT1 was highly homologous to winter rye CHT9, known to encode an AFP (Yeh et al., 2000). The objective of this study was to characterize a cold-inducible extracellular chitinase in bromegrass culture cells. Possible functions (chitinase activity and antifreeze activity) of BiCHT1 products were examined using transgenic E. coli and bromegrass cells. In addition, chemical and environmental factors affecting the expression of BiCHT1 were analyzed and compared with conditions for inducing antifreeze activity in the culture medium.
RESULTS
Deduced Amino Acid Sequence of BiCHT1
BiCHT1 cDNA was cloned from mRNA isolated from cold-treated bromegrass cells as previously described (Nakamura et al., unpublished data). The cDNA is 1,168 bp long with an open reading frame of 957 bp, encoding a peptide of 319 amino acids (predicted molecular mass is 33.2 kD). TargetP and PSORT analyses indicated with a certainty of 82% to 96.2% that there is a signal sequence of 20 amino acids that targets the protein to outside the cell (predicted molecular mass without the signal sequence is 31.3 kD). Comparison of the BiCHT1 amino acid sequence with known proteins demonstrated a homology with class I chitinase (Fig. 1). BiCHT1 protein had two consensus motifs (a chitin-binding domain and a catalytic domain) normally found in members of this family. BiCHT1 was highly homologous to winter rye CHT9, known as an AFP gene. The deduced amino acid sequence of BiCHT1 showed 89% identity with that of rye CHT9.
Figure 1.
Alignments of chitinase amino acid sequences deduced from nucleotide sequences of cDNAs from bromegrass BiCHT1 (accession no. AB428423), rye CHT9 (accession no. AF280437), wheat chi3 (accession no. AB029936), barley (accession no. U02287), and rice Cht-3 (accession no. D16223). Asterisks show conserved amino acid residues.
Proteins and Antifreeze Activity in the Culture Medium of Bromegrass during Cold Acclimation
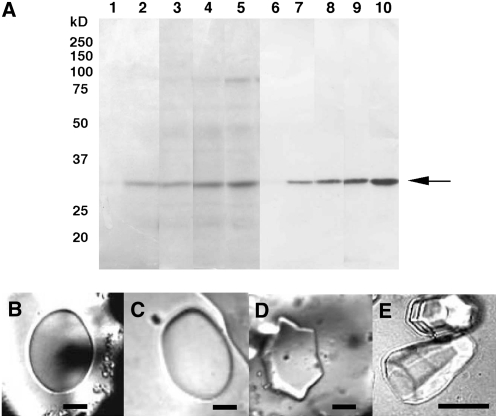
A 32-kD protein increased in the culture media of cold-acclimated wild-type bromegrass cells as detected by SDS-PAGE (Fig. 2A). The protein cross reacted with an antibody raised against rice (Oryza sativa) class I chitinase, CHT3. This allows us to conclude that the major protein increased in cold-acclimated bromegrass culture medium is a chitinase. The results coincided well with our microarray and RNA-blot data that showed that the expression of an endochitinase gene (BiCHT1) was up-regulated by cold and the mRNA accumulated with increasing exposure to cold (Nakamura et al., unpublished data).
Figure 2.
A, Coomassie Brilliant Blue-stained SDS-PAGE gels (lanes 1–5) and corresponding western blotting (lanes 6–10) of total proteins from culture media of bromegrass cells (wild type) exposed to low temperatures. Bromegrass cells were exposed to 15°C for a day (lanes 2, 7) and subsequently to 4°C for a day (lanes 3, 8), for 4 d (lanes 4, 9), and for 12 d (lanes 5, 10). Non-cold-treated samples were used as the control (lanes 1, 6). Twenty microliters of a 50-fold concentrated culture medium was applied to each lane. B to E, Induction of antifreeze activity in culture media of bromegrass exposed to low temperatures. Antifreeze activity was assayed by viewing the growth of an ice crystal formed in 50-fold concentrated culture medium. In these assays, the growth and morphology of ice crystals upon cooling were carefully observed. In the medium without antifreeze activity, flat round ice crystals grew rapidly. In the medium with antifreeze activity, ice crystal growth was retarded or arrested, and ice crystals showed specific shapes from flat hexagonal, hexagonal column to bipyramidal with increasing antifreeze activities (Hon et al. 1994). B, Before cold acclimation. C, At 15°C for a day then 4°C for a day. D, At 15°C for a day then 4°C for 5 d. E, At 4°C for 20 d. Bars = 50 μm.
No antifreeze activity or thermal hysteresis was detected in the culture medium before cold treatment, as a flat round ice crystal grew rapidly with decreasing temperature in the antifreeze assay (Fig. 2B). Antifreeze activity in the culture medium increased during cold acclimation as seen in the formation of a specific ice shape (hexagonal) and retardation of ice crystal growth after 5 d of treatment at 4°C (Fig. 2D). After 3 weeks of cold treatment, the ice crystal formed hexagonal column and bipyramidal shapes, indicating further increases in the antifreeze activity (Fig. 2E).
Expression of Recombinant BiCHT1 Protein in E. coli
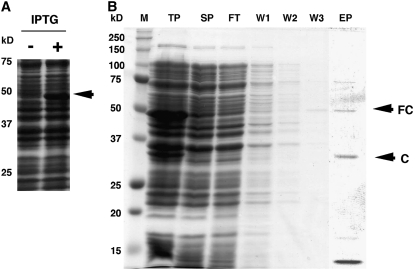
Expression and purification of recombinant BiCHT1 proteins were performed as described in “Materials and Methods.” As shown in Figure 3A, a tag-fused protein of 48 kD was detected by Coomassie Brilliant Blue staining after induction by isopropyl-β-d-thiogalactopyranoside (IPTG). Most recombinant BiCHT1 proteins were insoluble (recovered in the centrifugation pellet), but there were some soluble (recovered in the supernatant) recombinant proteins (Fig. 3B). The soluble, tag-fused protein bound to S-protein agarose and was successfully eluted after specific digestion with a protease to form a 33-kD recombinant protein (Fig. 3B). Both the tag-fused (48 kD) and tag-removed (33 kD) recombinant proteins cross reacted with anti-CHT3 antibody (data not shown). Chitinase activity was detected in the total soluble protein fraction containing the tag-fused protein and with a high specific activity in the tag-removed recombinant BiCHT1 protein fraction (Table I). However, the tag-removed recombinant BiCHT1 protein fraction had no antifreeze activity even at 2 mg/mL concentration (Fig. 6H).
Figure 3.
Coomassie Brilliant Blue-stained SDS-PAGE gels showing induction (A) and purification steps (B) of recombinant BiCHT1 expressed in E. coli carrying pET-BiCHT1 vector. An S·Tag-fused recombinant protein (48 kD, indicated by the arrow) induced by IPTG (A) was partially soluble (B; TP versus SP). The bacteria were sonicated in 5 mL of buffer (TP). Following centrifugation, the supernatant (SP, 5 mL) was applied to S-protein agarose, which binds only S·Tag-fused proteins. After three washes (W1–W3, 5 mL each), the S-protein agarose was incubated with a specific protease to release a tag-removed recombinant protein (EP, 5 mL). Each lane in SDS-PAGE represents 20 μL of each fraction. M, Molecular mass marker; TP, total proteins; SP, soluble proteins; FT, flow through; W1, wash 1; W2, wash 2; W3, wash 3; EP, eluted proteins; FC, tag-fused protein; C, tag-removed protein (chitinase).
Table I.
Specific activity of recombinant BiCHT1 proteins produced in E. coli and BiCHT1 products from transgenic bromegrass cell lines
Chitinase activity was measured as described in “Materials and Methods” and expressed as mean ± sd (n = 3).
| Proteins | Chitinase Activity |
|---|---|
| nmol min−1mg−1protein | |
| Total soluble proteins (pET32a) | <0.01 |
| Total soluble proteins (pET-BiCHT1; tag-fused) | 1.5 ± 0.1 |
| Recombinant BiCHT1 (tag-removed) | 92.5 ± 7.3 |
| Wild-type media | 10.7 ± 2.8 |
| T2 media | 13.4 ± 4.3 |
| T6 media | 17.8 ± 5.0 |
Figure 6.
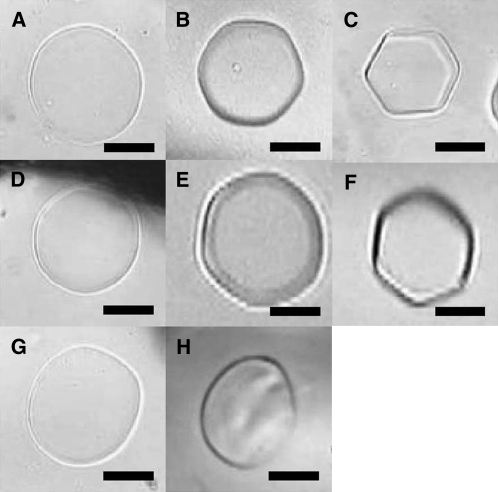
Antifreeze activity in the culture medium of wild-type bromegrass cells grown for 7 d at 25°C (A), 10°C (B), and 4°C (C) compared to antifreeze activity in the culture medium of transgenic bromegrass cell lines T6 grown for 7 d at 25°C (D), 10°C (E), and 4°C (F) and T2 grown for 7 d at 25°C (G). H, Antifreeze activity of recombinant BiCHT1 proteins (tag-removed). All the culture media were concentrated 50-fold for detection of antifreeze activity. Approximate concentrations of BiCHT1 proteins (mg/mL) were 0.1 (A), 0.14 (B), 0.04 (C), 0.35 (D), 0.3 (E), 0.2 (F), 0.25 (G), and 2.0 (H). Bars = 50 μm.
Overexpression of BiCHT1 Protein in Culture Media of Transgenic Bromegrass Cells and Antifreeze Activity of the Media
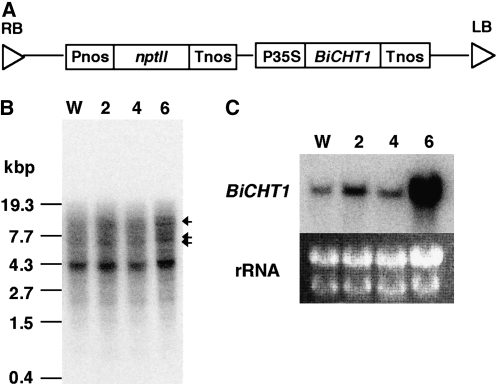
To investigate the native form of BiCHT1 protein, the BiCHT1 gene was introduced into bromegrass suspension-cultured cells under the control of a cauliflower mosaic virus (CaMV) 35S promoter (Fig. 4A). We obtained 19 geneticin-resistant bromegrass cell lines, in 12 lines of which transgene-specific PCR product was detected (data not shown). Southern-blot analysis (Fig. 4B) revealed that the introduced BiCHT1 gene was integrated into the genome in at least six of these transgenic bromegrass cell lines. The expression of BiCHT1 mRNA in the transformants was determined by northern-blot analysis (Fig. 4C). Two cell lines, one expressing relatively low levels of BiCHT1 mRNA (T2) and the other expressing high levels of BiCHT1 mRNA (T6), were selected for further experiments.
Figure 4.
Confirmation of the introduced gene (BiCHT1) and its expression. A, Map of the T-DNA region of the binary vector. B, Southern-blot analysis of transformants (cell lines T2, T4, and T6) and wild-type bromegrass cells (W). Arrows indicate the amplified copies of the introduced gene BiCHT1. C, Northern-blot analysis of BiCHT1 mRNA from transgenic lines (T2, T4, and T6) and wild-type cells (W) grown for 7 d at 25°C.
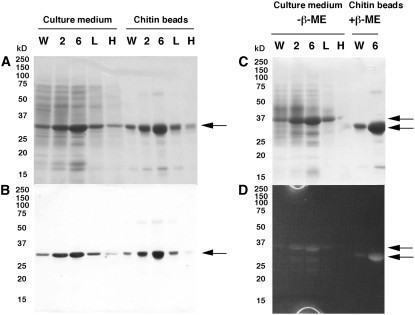
The culture medium of these transgenic lines showed increased chitinase activities compared to that of wild-type cells (Table I). SDS-PAGE of the culture medium revealed that two independent transgenic cell lines (T2 and T6) overexpressed the BiCHT1 products with the molecular mass identical to the one induced by exposure to 4°C and 10°C (Fig. 5A). The BiCHT1 products were confirmed to be purified from culture media by adsorption to chitin affinity beads (Fig. 5A). The molecular mass of the expressed protein (32 kD) was consistent with the calculated one (31.3 kD, without the signal sequence) of the BiCHT1 protein based on the deduced amino acid sequence. Western analysis showed cross reaction of the expressed proteins with anti-CHT3 antiserum (Fig. 5B). When SDS-PAGE of culture media was performed in the absence of β-mercaptoethanol (β-ME), the expressed BiCHT1 protein showed a decreased mobility (35 kD; Fig. 5C, top arrow) as compared to the mobility in the presence of β-ME (Fig. 5C, bottom arrow). Gel-based enzyme assays detected chitinase activities in both the 35-kD (−β-ME) and 32-kD (+β-ME) bands (Fig. 5D). This indicated that chitinase denatured in the presence of β-ME could refold in the gel to regain an active form during incubation with the buffer following the SDS-PAGE. These results allow us to conclude that the BiCHT1 product is a chitinase secreted into the culture medium.
Figure 5.
Coomassie Brilliant Blue-stained SDS-PAGE gels (A) showing overexpression of BiCHT1 protein (arrow) in the culture medium and its chitin-binding capability and the corresponding western-blot images using anti-CHT3 antibody (B). Coomassie Brilliant Blue-stained SDS-PAGE (C) performed in the presence or absence of β-ME and the corresponding zymogram analysis (in-gel detection of chitinase activity) images (D) of BiCHT1 protein in the culture medium or bound to chitin beads. W, Wild type (25°C); 2, 6, transformants T2, T6 (25°C); L, wild type (10°C); H, wild type (4°C). All samples were grown for 7 d at the designed temperatures (the cells were directly incubated at 4°C or 10°C, which resulted in lower accumulation of BiCHT1 product at 4°C than shown in Fig. 2 because direct exposure to 4°C retarded cell growth). Twenty microliters of a 50-fold concentrated culture medium or an equivalent was loaded onto each lane.
However, antifreeze activity was not detected in the culture medium (50-fold concentrated) of transgenic bromegrass cell lines (T6 and T2) grown for 7 d at 25°C (Fig. 6, D and G). These samples contained approximately 0.25 to 0.35 mg/mL of the BiCHT1 product. Further concentration of the culture media to 200-fold failed to show antifreeze activity (data not shown). Antifreeze activity was not detected even after the addition of 5 to 20 mm CaCl2, KCl, NaCl, or EDTA (data not shown). Preincubation of the harvested culture medium (50-fold concentrated, molecular weight [MW] > 5 kD) at 4°C for 1 to 7 d in the presence and absence of medium filtrates (MW cutoff <5 kD) from wild-type cell cultures grown at 4°C for 3 weeks did not induce antifreeze activity (data not shown). In the meantime, higher antifreeze activity was detected in the 50-fold concentrated culture medium from wild-type cells grown at 4°C (ice crystals with sharper hexagonal shapes formed) than from those grown at 10°C (Fig. 6, C compared to B). The levels of the BiCHT1 protein in these samples were approximately 0.04 mg/mL (4°C) and 0.14 mg/mL (10°C). Antifreeze activity levels seemed unrelated with BiCHT1 protein concentrations.
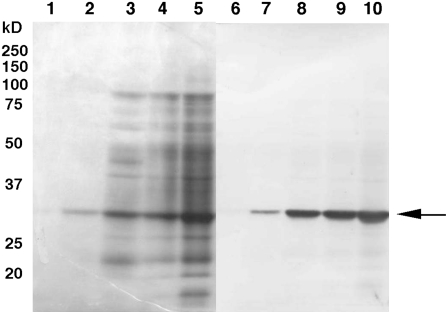
When the transgenic bromegrass cell line (T6) was grown at 4°C and 10°C for 7 d, the culture medium (50-fold concentrated) showed antifreeze activity, but the levels were similar to those of wild-type cells: wild type at 4°C = T6 at 4°C > wild type at 10°C = T6 at 10°C (Fig. 6, F compared to C, and E compared to B). Serial dilution of these samples showed similar declines in the activity (data not shown). SDS-PAGE and western-blot analyses revealed that the levels of BiCHT1 product in these samples were: T6 at 10°C > T6 at 4°C ≫ wild type at 10°C > wild type at 4°C (Fig. 7). BiCHT1 protein levels of these samples seemed unrelated with antifreeze activity levels.
Figure 7.
Coomassie Brilliant Blue-stained SDS-PAGE (lanes 1–4) and corresponding western-blot images (lanes 5–8) showing differential expression of BiCHT1 protein (arrow) in the medium of cold-acclimated wild-type and transgenic bromegrass cultures. Wild-type bromegrass cultures were grown at 4°C (lanes 1 and 5) and 10°C (lanes 2 and 6) for 7 d. Transgenic bromegrass cultures (T6) were similarly grown at 4°C (lanes 3 and 7) and 10°C (lanes 4 and 8) for 7 d. Twenty microliters of a 50-fold concentrated culture medium was applied to each lane.
Expression of BiCHT1 Proteins during a Normal Culture Condition
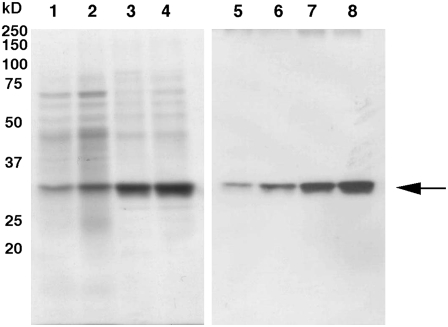
The expression of BiCHT1 chitinase was investigated in the media of wild-type bromegrass suspension cells at a normal (no stress/no treatment) growth condition by SDS-PAGE and western blots (Fig. 8). The chitinase protein in the culture media gradually increased during growth at 25°C, with the highest accumulation on 10- to 14-d incubation periods, which correspond to the stationary phase in the culture growth stage (Ishikawa et al., 2006; Nakamura and Ishikawa, 2006).
Figure 8.
Expression of BiCHT1 proteins in the culture medium of wild-type bromegrass cells grown at 25°C. Cultures were initiated by inoculating 1.5 g of wild-type bromegrass cells. Coomassie Brilliant Blue-stained SDS-PAGE gels (lanes 1–5) showing expression of BiCHT1 protein (arrowed) in the culture medium and their corresponding western-blot images using anti-CHT3 antibody (lanes 6–10). Culture media were prepared from cultures incubated for a day (lanes 1, 6), 3 d (lanes 2, 7), 7 d (lanes 3, 8), 10 d (lanes 4, 9), and 14 d (lanes 5, 10) after inoculation. Twenty microliters of a 50-fold concentrated culture medium was applied to each lane.
Effect of Various Stress, Hormone, and Chemical Treatments on BiCHT1 Expression and Antifreeze Activity
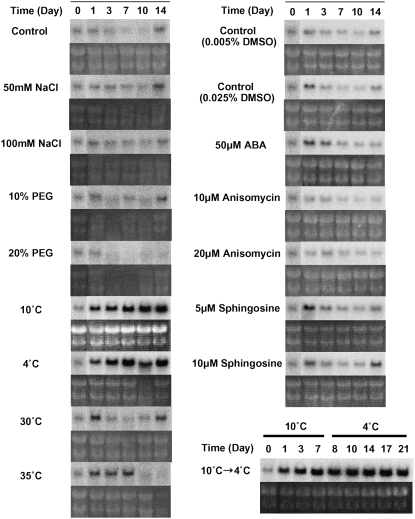
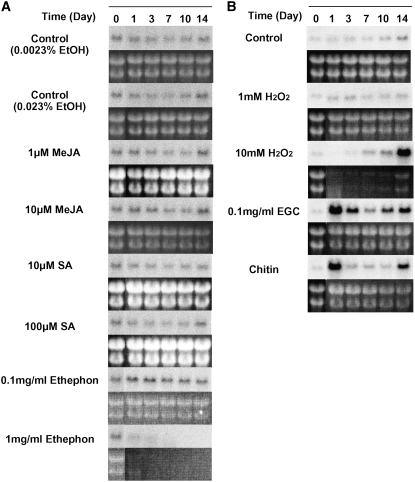
Northern-blot analyses were performed to determine how the expression of the BiCHT1 gene is regulated in response to environmental stresses. The data in Figure 9 shows that the levels of BiCHT1 mRNA were slightly increased at the stationary phase (14 d after inoculation). The expression of BiCHT1 was most increased by incubation at 4°C and 10°C and moderately at 35°C. BiCHT1 mRNA was not increased by salt or water stress. Abscisic acid (ABA), anisomycin, and sphingosine have been shown to induce freezing tolerance in bromegrass cultured cells at nonhardening temperatures (Ishikawa et al., 1990; Wu et al., 2004) but failed to increase the expression of BiCHT1 mRNA compared to the respective controls (dimethyl sulfoxide [DMSO] only).
Figure 9.
Northern-blot analyses showing the expression of BiCHT1 mRNA in wild-type bromegrass culture cells grown for 0 to 14 d under various stress and chemical treatments (the top segment in each pair). Ribosomal RNA was visualized by ethidium bromide staining (the bottom segment in each pair). Anisomycin and d-erythro-sphingosine were dissolved in DMSO and were added to the culture medium.
In Figure 10, methyl jasmonate (MeJA), salicylic acid (SA), and ethylene (given in the form of ethephon), which are known to be involved in pathogenesis-related responses, did not increase the expression of BiCHT1 mRNA as compared to the respective controls (ethanol only). The expression of BiCHT1 mRNA was rapidly but only transiently increased 7- to 9-fold by the addition of soluble chitin (ethylene glycol chitin [EGC]) and insoluble chitin. Incubating cells with 1 mm hydrogen peroxide (H2O2) did not affect the expression. Prolonged exposure (14 d) to 10 mm H2O2 and chitin (soluble or insoluble) increased the mRNA level in a manner that intensified the stationary phase-specific increase of the expression.
Figure 10.
Northern-blot analyses showing the expression of BiCHT1 mRNA in wild-type bromegrass culture cells grown for 0 to 14 d under various treatments related to pathogenesis (the top segment in each pair). Ribosomal RNA was visualized by ethidium bromide staining (the bottom segment in each pair). A, Treated with plant hormones. B, Treated with elicitors. MeJA, SA, and ethephon were dissolved in ethanol and were added to the culture medium.
Antifreeze activity in culture media (50-fold concentrated) of cells grown with some of these treatments was determined (data not presented). No antifreeze activity was detected in the medium of cells grown at 25°C in the presence of 75 μm ABA for 7 to 14 d or in the presence of chitin for 7 d. Prolonged incubation (14 d) of cells at 25°C without any treatment did not induce antifreeze activity in the medium. Antifreeze activity was detected in the medium concentrates of cells grown with ethephon (0.7–7 mm) at 25°C for 2 and 4 d.
DISCUSSION
Freezing is one of the most critical environmental stresses, and freezing tolerance is important for plants to survive severe cold winter periods. Plants show diverse levels of adaptation to cold (Thomashow, 1998), and adaptations involve regulation of enzyme activities and changes in gene expression. Smooth bromegrass is one of the most cold hardy grass species (Sakai and Larcher, 1987) and may serve as an important source of freezing tolerance genes.
Freezing tolerance is induced in bromegrass suspension cells by exposure to low temperatures (Robertson et al., 1987; Ishikawa et al., 1990). To understand the mechanisms of cold acclimation and freezing tolerance in this system, the genes responding to low temperatures were investigated. Using the microarray technique, we have previously cloned several genes from bromegrass (Nakamura et al., unpublished data). In this report, we describe the characterization of a cold-responsive transcript, BiCHT1, encoding a putative chitinase, a member of the glycoside hydrolase family 19 chitinase. BiCHT1 was highly homologous to winter rye CHT9, known to encode an AFP. Transcripts of BiCHT1 accumulated with increasing periods of cold acclimation (4°C and 10°C) in bromegrass cells (Nakamura et al., unpublished data). The same tendency was observed at the protein level (Fig. 2A).
Expression of BiCHT1 products in E. coli was difficult. Recombinant BiCHT1 proteins expressed in E. coli BL21 (DE3) pLysS were completely aggregated and insoluble (data not shown). On the other hand, E. coli Origami (DE3) pLysS, which has oxidative cytoplasm, could partially produce recombinant BiCHT1 proteins in a soluble form (Fig. 3B). Therefore, formation of sufficient disulfide bonds may be required for solubilization of recombinant BiCHT1 proteins. ScanProsite analysis predicted the presence of four disulfide bonds in the chitin-binding domain and its vicinity, which may be important for the chitin-binding activity. The soluble BiCHT1 products obtained (tag-fused, tag-removed) had chitinase activity (Table I), but no antifreeze activity was detected in these protein fractions (BiCHT1 product concentration in the tag-removed fraction was 2 mg/mL; Fig. 6H).
Protein structure analysis showed that BiCHT1 has a chitin-binding domain and a catalytic domain and was expected to be secreted. The BiCHT1 gene was successfully introduced by Agrobacterium-mediated transformation into bromegrass cells and overexpressed using the CaMV 35S promoter. There was an increase in a 32-kD protein band in SDS-PAGE gels of total protein fractions from the culture medium of overexpressing transgenic lines (Fig. 5). This band cross reacted with anti-CHT3 antibody and also bound to chitin beads. The culture medium of overexpressing lines had increased chitinase activities (Table I). The enzymic activities were subjected to zymogram analyses. Zymograms are not quantitative; however, they do offer a rough estimate of the approximate molecular masses of active enzymes. The chitinase zymogram (Fig. 5D) revealed one band with chitinolytic activity with an approximate molecular mass of 32 kD. These data confirmed the production and secretion of BiCHT1 proteins with the proper chitinase function (most probably with the proper structure) by the transformed bromegrass cells. However, the culture medium concentrates of BiCHT1 overexpressing cell lines did not modify the growth of ice in the antifreeze assays (BiCHT1 product concentration, 0.25–0.35 mg/mL; Fig. 6, D and G). Antifreeze activities were not detected even at the concentration of 1.0 to 1.4 mg/mL. At the concentration (0.25–2 mg/mL) used for assaying the products from transgenic E. coli and bromegrass, apoplast chitinases isolated from cold-acclimated winter rye showed high antifreeze activities (bipyramid ice crystals) according to Hon et al. (1994, 1995). Carrot (Daucus carota) antifreeze protein is also known to show high thermal hysteresis at this concentration range (D.-Q. Zhang et al., 2004).
Antifreeze activity was detected in the apoplast of freezing-tolerant cereals (Antikainen and Griffith, 1997). Antifreeze activity was detected in the culture medium of bromegrass suspension cultures grown at 4°C (Fig. 2). Seemingly, the expression pattern of BiCHT1 products at 4°C resembled that of antifreeze activities. However, BiCHT1 products expressed in both E. coli and bromegrass had chitinase activity but did not affect the growth of ice crystals. Stressmann et al. (2004) reported that when winter rye apoplast crude extract containing chitinase/AFP was frozen in the presence of 20 to 200 mm of CaCl2, the antifreeze activity was reduced by 30% and was recovered by adding a chelator. This effect was specific to high concentrations (20–200 mm) of CaCl2, and antifreeze activity before freeze-thaw cycles was unaffected by the presence of CaCl2. In our antifreeze assays, freshly harvested samples were immediately used for assays without any freeze-thaw processes. Erickson's (ER) medium contains 0.1 mm of EDTA and 3 mm of CaCl2, and after 1 to 2 weeks of culture, the Ca concentration was 1 to 2 mm, which is much lower than the concentrations reported to be effective. Therefore, the possible Ca effect on antifreeze activity in our system is considered to be minimal. In fact, antifreeze activities were well detected in the culture medium of bromegrass cells incubated at 4°C. Type II AFPs of fish are known to require Ca2+ for the antifreeze activity (Ewart et al., 1999). To double-check if the activity interacts similarly with cations, the BiCHT1 product was examined for antifreeze activity in the presence of various concentrations of CaCl2, other cations, and chelators. The BiCHT1 product, after harvest, was also incubated at 4°C for 1 to 7 d before antifreeze assays to promote any possible protein refolding. However, antifreeze activity was not detected nor recovered under any of these conditions tested (data not shown).
When the transgenic bromegrass cell lines were grown at 4°C, the culture medium concentrates showed antifreeze activity levels similar to that of wild-type cells grown at 4°C (Fig. 6) in spite of greater accumulation of BiCHT1 products in the medium (Fig. 7). This allows us to discount the possibility that some cofactors synthesized during cold acclimation at 4°C may be required for the latent antifreeze activity of the BiCHT1 product to become detectable. We also found that antifreeze activity in the medium concentrates of cells grown at 4°C was not affected by the addition of anti-CHT3 antibody (data not shown). This implies that the protein domain responsible for the antifreeze activity was not affected by binding of anti-CHT3 antibody.
Numerous types of AFP have been isolated from different organisms. A common structural feature of many animal AFPs is the presence of repeated amino acid sequences. Repeat sequences have been found in fish antifreeze glycoproteins, fish type I and type IV AFPs, and insect AFPs, while fish type II and III AFPs have no repetitive sequences (Cheng, 1998). In plant AFPs, clear repeated sequences have been found in carrot, Lolium, and Solanum AFPs, but not in winter rye AFPs (Cheng, 1998; Worrall et al., 1998; Meyer et al., 1999; Huang and Duman, 2002). Structural studies of winter flounder type I AFP (Sicheri and Yang, 1995), beetle AFP (Liou et al., 2000), Lolium AFP (Kuiper et al., 2001), and carrot AFP (D.-Q. Zhang et al., 2004) indicate that the proper repeated sequence is necessary to provide a spatial match between the binding groups of the AFPs and water molecules to confer ice-binding ability via hydrogen bonding. Primary structure analyses using Radar, REP, and REPRO in ExPASy Proteomics tools (http://us.expasy.org/tools/) revealed that there are no distinct repetitive amino acid sequences in the BiCHT1 product (results not shown).
BiCHT1 is a single-copy gene in bromegrass (data not shown). The antibody used in the western-blot studies specifically cross reacts with the Cht-3 product and not with Cht-2 in the class I chitinase family of rice (Nishizawa et al., 1999). Taken together, it is unlikely that the bands recognized by the antibody represent chitinases of similar size other than the BiCHT1 product (Figs. 2, 5, 7, and 8).
Expression of BICHT1 was up-regulated and its product accumulated in long-term exposure to low temperatures (Figs. 2, 5, 7, and 9). The level of BiCHT1 mRNA expression at 10°C was similar to that at 4°C (Fig. 9) in contrast to antifreeze activity levels described below. It was also expressed at 35°C. Many genes induced by temperature stresses are also known to be induced by salt or drought stress (Seki et al., 2003). Abiotic stresses such as NaCl and polyethylene glycol treatments did not induce the accumulation of BiCHT1 mRNA. ABA and ethephon treatment marginally influenced the expression of BiCHT1. Prolonged incubation at 25°C also moderately increased BiCHT1 mRNA and its product (Figs. 9 and 10). Chitinase induction is often regulated in response to pathogen attack, as well as in response to treatment with MeJA, SA, and elicitors (Kasprzewska, 2003). Expression of rice Cht-1 and Cht-3, one of the BiCHT1 homologs (Fig. 1), was induced by chitin or SA (Nishizawa and Hibi, 1991; Nishizawa et al., 1993). The expression of BiCHT1 was increased by chitin and a high concentration of H2O2, but was not up-regulated by MeJA or SA. Regulation of BiCHT1 expression seems different from other known chitinases.
Antifreeze activity detected in the culture medium was induced by incubation of bromegrass cells at 4°C but only slightly at 10°C (Fig. 6, C and B). It was also induced by ethephon (0.7–7 mm) treatment (data not shown). But it was not induced by ABA (50–75 μm) treatment nor by chitin or long-term incubation (14 d) at 25°C (data not shown). Antifreeze activity detected in the apoplasts of winter rye plants has been known to be induced by cold treatment, drought (Yu and Griffith, 2001), and ethylene (Yu et al., 2001) but not by ABA, SA, or snow molds (Hiilovaara-Teijo et al., 1999; Yu and Griffith, 2001). Although plant species (rye versus bromegrass) and systems used (leaves versus suspension culture) were different, our observations on the regulation of antifreeze activity are consistent to Griffith and Yaish (2004). It was also noted that the bromegrass antifreeze activity was induced by different stimuli and conditions (4°C, ethephon) than those stimulating the expression of BiCHT1 mRNA (4°C, 10°C, 35°C, chitin, prolonged incubation at 25°C, etc.; Figs. 9 and 10). When we focus on cold stress, the expression and accumulation levels of BiCHT1 mRNA per unit sampled cells were similar at 4°C and 10°C (Fig. 9). At the protein level, there was more accumulation of BiCHT1 products per unit cell culture medium at 10°C than at 4°C (Figs. 5 and 7). This was mostly due to the differences in cell growth at these temperatures; the growth was retarded but still greater at 10°C than at 4°C. In contrast, the level of antifreeze activity per unit culture medium was much higher at 4°C than at 10°C (Fig. 6, B and C), which was contrary to the levels of BiCHT1 protein in the medium. Moreover, transgenic cultures grown at 4°C and 10°C showed antifreeze activity similar to that of respective wild-type cultures in spite of much higher BiCHT1 product levels in the medium (Figs. 6 and 7). There seemed to be no correlation between antifreeze activity levels and BiCHT1 protein concentrations.
The results of transgenic E. coli and bromegrass cells and expression analyses of BiCHT1, taken together, allow us to conclude that the BiCHT1 product is a major protein with chitinase activity secreted in the medium from cold-treated bromegrass cells but is unlikely to be responsible for cold-induced antifreeze activity in the culture medium by itself, although BiCHT1 is highly homologous to winter rye CHT9, known as an AFP gene. Isolation of alternative AFP candidates from bromegrass is currently under way in our laboratory.
We are currently investigating possible functions of BiCHT1 and its product using the bromegrass cell lines overexpressing BiCHT1. Cold-inducible wheat chitinases, one of which has high homology to BiCHT1 and rye CHT9, have recently been found to be involved in snow mold resistance (Kawakami and Terami, 2005). Because the BiCHT1 product has chitinase activity and is induced by cold and chitin, similar functions may be expected. The expression of BiCHT1 is most pronounced under cold treatment and it is also expressed when the cells are in the stationary phase. The protein was expressed at 35°C, which is close to the threshold lethal temperature for bromegrass cells. These properties allow us to speculate that the product may possibly be involved in cellular metabolism or protection when cell culture growth is retarded.
MATERIALS AND METHODS
Plant Material and Culture Conditions
A nonembryogenic suspension culture of smooth bromegrass (Bromus inermis) ‘Manchar’ was used in this work, and growth conditions were as described previously (Nakamura and Ishikawa, 2006). Briefly, cultures were subcultured biweekly by transferring 5 mL of cells to 50 mL of fresh ER medium (Gamborg and Wetter, 1975), pH 5.8 (0.5 mg/L 2,4-dichlorophenoxyacetic acid) and incubated on a rotary shaker (85 rpm) at 25°C. For various treatments, cultures were initiated by inoculating 1.5 g of cells as described below.
Treatment of Cultured Cells with Stresses, Chemicals, and Elicitors
Bromegrass cells were subjected to salt stress by adding NaCl to the culture medium at a final concentration of 50 mm or 100 mm. Water stress was provided by adding 100 g/L or 200 g/L of polyethylene glycol to the culture medium. Cold treatment was conducted by transferring cultures grown for 3 d at 25°C either directly to 10°C or 4°C (Figs. 5–7) or following 1 d acclimation at 15°C (Fig. 2). Heat treatment was conducted by exposing 3-d-old cultures (grown at 25°C) at 30°C or 35°C. Long-term incubation at 35°C was lethal to bromegrass cells.
For ABA treatment, (±) ABA (Sigma-Aldrich) was added to the ER medium at a final concentration of 50 μm. Anisomycin (in DMSO) was added to cell cultures at a final concentration of 10 μm or 20 μm. d-Erythro-sphingosine (in DMSO) was added to cell cultures at a final concentration of 5 μm or 10 μm. A 10 mm MeJA stock solution (in ethanol) was added to cell cultures at a final concentration of 1 μm or 10 μm. A 100 mm SA stock solution (in ethanol) was added to cell cultures at a final concentration of 10 μm or 100 μm. A 100 mg/mL ethephon stock solution (in ethanol) was added to cell cultures at a final concentration of 0.1 mg/mL (0.7 mm) or 1 mg/mL (7 mm). However, prolonged incubation with 1 mg/mL ethephon was sometimes lethal to bromegrass cells. H2O2 solution was added to cell cultures at a final concentration of 1 mm or 10 mm. For chitin treatment, 5 mg of EGC (soluble) or 30 mg of chitin (insoluble) was added to 50 mL cell culture medium. All these chemical, hormone, and stress treatments were performed at 25°C. The cells were harvested at designated time periods during stress treatments or incubation with the chemicals and used for total RNA extraction.
Preparation of Proteins from Culture Media
Bromegrass cell cultures were filtered with an 80-μm nylon mesh, and the obtained liquid media were filtered to remove cell debris using a 0.22-μm Millex-GP syringe-driven filter (Millipore). The media were concentrated using Apollo 20 mL (QMWL, 5 kD; Orbital Biosciences) for a 50-fold concentration of extracellular proteins. The concentrated media was used for SDS-PAGE, western-blot analyses, and antifreeze activity assays.
Assay of Antifreeze Activity
Antifreeze activity was detected according to Hon et al. (1994) using a nanoliter osmometer (Clifton Technical Physics) assembled on an Olympus microscope (BX50). In the absence of antifreeze activity, ice crystals grow to form a flat round disc. Ice crystals in a solution having antifreeze activity grow in a retarded manner to form hexagonally shaped crystals or bipyramidal crystals. Ice crystal growth was monitored with a video camera (CS580, Olympus) and recorded as video files in the computer. When necessary, antifreeze activity was measured in the presence of various cations (5–20 mm CaCl2, KCl, MgCl2, and NaCl), chelators (5–20 mm EDTA), or anti-CHT3 antibody (50-fold diluted) to check if these conditions affected the observed antifreeze activities. Antifreeze activity was also determined with 50-fold concentrated media from cultures grown in the presence of ABA (50–75 μm), ethephon (0.7–7 mm), or insoluble chitin (30 mg in 50 mL) at 25°C for 2 to 14 d besides cold-treated cultures. In some cases, culture media (50-fold concentrated, MW > 5 kD) harvested from transgenic cultures (grown at 25°C for 7–14 d) were incubated at 4°C for 1 to 7 d in the presence and absence of medium filtrates (MW cutoff <5 kD) from wild-type cell cultures grown at 4°C for 3 weeks prior to antifreeze assays.
Northern- and Southern-Blot Analyses
Northern- and Southern-blot analyses were performed as described previously (Nakamura et al., 1997). Partial BiCHT1 cDNA (1–872 base) including the 5′ noncoding region was used as a probe. For Southern-blot analysis, 20 μg of genomic DNA from three selected transgenic bromegrass cell lines and a nontransformed line was digested with HindIII.
Expression of BiCHT1 in Escherichia coli
For construction of the chitinase expression vector, PCR was performed using BiCHT1 cDNA as a template with the forward primer 5′-GCCACCATGGGAGGACTTGTGGTGGCG-3′ and reverse primer 5′-TCATTACTATGCGAACGGCCTCTG-3′. The PCR products were cloned into the pGEM-T Easy vector (Promega). After confirmation of the DNA sequence, the DNA fragment encoding BiCHT1 was excised by digestion with NcoI and EcoRI, and ligated into the expression vector pET-32a (Novagen), previously digested by the same enzymes. The recombinant plasmid, pET-BiCHT1, was introduced into E. coli Origami (DE3) pLysS (Novagen). For induction of recombinant BiCHT1 protein, 50-mL suspension (cell density, A600 = 1.0) of E. coli harboring pET-BiCHT1 was incubated with 1 mm IPTG for 24 h at 20°C. The bacteria harvested by centrifugation were sonicated in 5 mL of buffer comprising 50 mm Tris-HCl, pH 8.0, 50 mm NaCl, 1 mm EDTA, and centrifuged to obtain the supernatant (soluble protein fraction). Recombinant BiCHT1 was purified from the soluble protein fraction (5 mL) using an S·Tag rEK Purification kit (Novagen) according to the method specified by the supplier.
SDS-PAGE and Western-Blot Analysis
SDS-PAGE was carried out by the method of Laemmli (1970) using 12.5% polyacrylamide gels and Coomassie Brilliant Blue staining unless otherwise noted. Western-blot analysis was performed using an anti-chitinase mouse antibody raised against recombinant rice (Oryza sativa) CHT3 (Nishizawa et al., 1999). The primary antibodies were detected with secondary anti-mouse IgG conjugated with alkaline phosphatase to generate a nitro blue tetrazolium chloride/5-bromo-4-chloro-3′-indolylphosphatase p-toluidine salt signal. The concentrations of BiCHT1 products were estimated from the intensity of the responsible bands in western blots as compared to those of known amounts of chitinase run on the same gel.
Chitinase Activity
Chitinase activity was assayed colorimetrically with glycol chitin as a substrate. The sample solution (100 μL) was added to 900 μL of 100 mm sodium acetate buffer, pH 7.0, containing 0.1% (w/v) EGC (Seikagaku). After incubation of the reaction mixture at 37°C for 5 h, the amount of reducing sugars produced was determined by reading A420 using the modified Schales method (Imoto and Yagishita, 1971).
To examine chitin-binding ability, a slurry of 500 μL of chitin beads (New England Biolabs) was added to 50 mL of culture medium. After gentle agitation at 4°C for 2 h, the beads were pelleted and washed extensively with 100 mm sodium phosphate buffer, pH 7.0. The chitin beads were resuspended in 200 μL of SDS-PAGE loading buffer and boiled for 5 min. The samples were directly used for SDS-PAGE analysis.
For zymogram assays of chitinase activity, samples were analyzed on 12.5% SDS-polyacrylamide gels containing 0.01% (w/v) EGC according to Trudel and Asselin (1989). After electrophoretic separation, the gel was incubated with agitation at 37°C for 2 h in 100 mm sodium acetate buffer, pH 5.0, containing 1% (v/v) Triton X-100, and subsequently incubated for 10 min at room temperature in a staining solution (500 mm Tris-HCl, pH 8.9, 0.01% [w/v] Fluorescent Brightener 28 [Sigma-Aldrich]). Then the gel was destained for 1 h in distilled water. Lytic zones were visualized under UV light as nonfluorescent bands.
Transformation of Bromegrass Culture Cells
BiCHT1 cDNA was cloned into a binary vector, pBI121, under the control of a CaMV 35S promoter. The resultant plasmid, pBI-BiCHT1 (Fig. 4A), was introduced into Agrobacterium tumefaciens EHA105 using a freeze-thaw method (Holsters et al., 1978). Transformation of bromegrass cultured cells was achieved by cocultivation with Agrobacterium and selection for geneticin resistance as described previously (Nakamura and Ishikawa, 2006).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AB428423 (BiCHT1).
Acknowledgments
We thank Dr. Y. Nishizawa (NIAS) for the gift of anti-chitinase antibody and Ms Y. Koike and R. Iwanami (NIAS) for maintenance of the cultures. We thank Dr. A.J. Robertson (University of Saskatchewan) for critical reading of the manuscript. We dedicate this article to the late Prof. Marilyn Griffith who made great contributions to antifreeze studies in plants with her pioneering works and also kindly showed us antifreeze assays.
This work was supported by the Rice Genome Project (grant nos. MA–2121 and PR1207); by the Bio Design Program from Ministry of Agriculture, Forestry and Fisheries, Japan; by Bio-oriented Technology Research Advancement Institution, Japan (a PRBRAIB grant); and by Japan Society for the Promotion of Science (grants-in-aid for scientific research nos. 17658011 and 16380030 to M.I.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Masaya Ishikawa (isikawam@affrc.go.jp).
Open Access articles can be viewed online without a subscription.
References
- Antikainen M, Griffith M (1997) Antifreeze protein accumulation in freezing-tolerant cereals. Physiol Plant 99 423–432 [Google Scholar]
- Bohnert HJ, Nelson DE, Jensen RG (1995) Adaptation to environmental stress. Plant Cell 7 1099–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C-HC (1998) Evolution of the diverse antifreeze proteins. Curr Opin Genet Dev 8 715–720 [DOI] [PubMed] [Google Scholar]
- Chun JU, Yu XM, Griffith M (1998) Genetic studies of antifreeze proteins and their correlation with winter survival in wheat. Euphytica 102 219–226 [Google Scholar]
- De Jong AJ, Cordewener J, Lo Shiavo F, Terzi M, Vandekerckhove J, Van Kammen A, De Vries SC (1992) A carrot somatic embryo mutant is rescued by chitinase. Plant Cell 4 425–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewart KV, Lin Q, Hew CL (1999) Structure, function and evolution of antifreeze proteins. Cell Mol Life Sci 55 271–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamborg OL, Wetter LR (1975) Plant Tissue Culture Methods. National Research Council of Canada, Saskatoon, Canada
- Griffith M, Yaish MW (2004) Antifreeze proteins in overwintering plants: a tale of two activities. Trends Plant Sci 9 399–405 [DOI] [PubMed] [Google Scholar]
- Guy CL (1990) Cold acclimation and freezing stress tolerance: role of protein metabolism. Annu Rev Plant Physiol Plant Mol Biol 41 187–223 [Google Scholar]
- Hiilovaara-Teijo M, Hannukkala A, Griffith M, Yu XM, Pihakaski-Maunsbach K (1999) Snow-mold-induced apoplastic proteins in winter rye leaves lack antifreeze activity. Plant Physiol 121 665–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsters M, De Waele D, Depicker A, Messens E, Van Montagu M, Schell J (1978) Transfection and transformation of Agrobacterium tumefaciens. Mol Gen Genet 163 181–187 [DOI] [PubMed] [Google Scholar]
- Hon WC, Griffith M, Chong P, Yang DSC (1994) Extraction and isolation of antifreeze proteins from winter rye (Secale cereale L.) leaves. Plant Physiol 104 971–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hon WC, Griffith M, Mlynarz A, Kwok YC, Yang DSC (1995) Antifreeze proteins in winter rye are similar to pathogenesis-related proteins. Plant Physiol 109 879–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T, Duman JG (2002) Cloning and characterization of a thermal hysteresis (antifreeze) protein with DNA-binding activity from winter bittersweet nightshade, Solanum dulcamara. Plant Mol Biol 48 339–350 [DOI] [PubMed] [Google Scholar]
- Imoto T, Yagishita K (1971) A simple activity measurement of lysozyme. Agric Biol Chem 35 1154–1156 [Google Scholar]
- Ishikawa M, Robertson AJ, Gusta LV (1990) Effect of temperature, light, nutrients and dehardening on abscisic acid induced cold hardiness in Bromus inermis Leyss suspension cultured cells. Plant Cell Physiol 31 51–59 [Google Scholar]
- Ishikawa M, Suzuki M, Nakamura T, Kishimoto T, Robertson AJ, Gusta LV (2006) Effect of growth phase on survival of bromegrass suspension cells following cryopreservation and abiotic stresses. Ann Bot (Lond) 97 453–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasprzewska A (2003) Plant chitinases: regulation and function. Cell Mol Biol Lett 8 809–824 [PubMed] [Google Scholar]
- Kawakami A, Terami F, inventors. January 4, 2005. Low temperature expression chitinase cDNAs and method for isolating the same. U.S. Patent Application No. 6838271
- Kuiper MJ, Davies PL, Walker VK (2001) A theoretical model of a plant antifreeze protein from Lolium perenne. Biophys J 81 3560–3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685 [DOI] [PubMed] [Google Scholar]
- Liou YC, Tocilj A, Davies PL, Jia Z (2000) Mimicry of ice structure by surface hydroxyls and water of a β-helix antifreeze protein. Nature 406 322–324 [DOI] [PubMed] [Google Scholar]
- Meyer K, Keil M, Naldrett MJ (1999) A leucine rich repeat protein of carrot that exhibits antifreeze activity. FEBS Lett 447 171–178 [DOI] [PubMed] [Google Scholar]
- Mouille G, Robin S, Lecomte M, Pagant S, Höfte H (2003) Classification and identification of Arabidopsis cell wall mutants using Fourier Transform InfraRed (FT-IR) microspectroscopy. Plant J 35 393–404 [DOI] [PubMed] [Google Scholar]
- Nakamura T, Ishikawa M (2006) Transformation of suspension cultures of bromegrass (Bromus inermis Leyss) by Agrobacterium tumefaciens. Plant Cell Tissue Organ Cult 84 293–299 [Google Scholar]
- Nakamura T, Yokota S, Muramoto Y, Tsutsui K, Oguri Y, Fukui K, Takabe T (1997) Expression of a betaine aldehyde dehydrogenase gene in rice, a glycinebetaine nonaccumulator, and possible localization of its protein in peroxisomes. Plant J 11 1115–1120 [DOI] [PubMed] [Google Scholar]
- Nishizawa Y, Hibi T (1991) Rice chitinase gene: cDNA cloning and stress-induced expression. Plant Sci 76 211–218 [Google Scholar]
- Nishizawa Y, Kishimoto N, Saito A, Hibi T (1993) Sequence variation, differential expression and chromosomal location of rice chitinase genes. Mol Gen Genet 241 1–10 [DOI] [PubMed] [Google Scholar]
- Nishizawa Y, Nishio Z, Nakazono K, Soma M, Nakajima E, Ugaki M, Hibi T (1999) Enhanced resistance to blast (Magnaporthe grisea) in transgenic rice by constitutive expression of rice chitinase. Theor Appl Genet 99 383–390 [DOI] [PubMed] [Google Scholar]
- Robertson AJ, Gusta LV, Reaney MJT, Ishikawa M (1987) Protein synthesis in bromegrass (Bromus inermis Leyss.) cultured cells during the induction of frost tolerance by abscisic acid or low temperature. Plant Physiol 84 1331–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai A, Larcher W (1987) Frost Survival of Plants. Springer-Verlag, Berlin
- Seki M, Kamei A, Yamaguchi-Shinozaki K, Shinozaki K (2003) Molecular responses to drought, salinity and frost: common and different paths for plant protection. Curr Opin Biotechnol 14 194–199 [DOI] [PubMed] [Google Scholar]
- Sicheri F, Yang DS (1995) Ice-binding structure and mechanism of an antifreeze protein from winter flounder. Nature 375 427–431 [DOI] [PubMed] [Google Scholar]
- Stressmann M, Kitao S, Griffith M, Moresoli C, Bravo LA, Marangoni AG (2004) Calcium interacts with antifreeze proteins and chitinase from cold-acclimated winter rye. Plant Physiol 135 364–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF (1998) Role of cold-responsive genes in plant freezing tolerance. Plant Physiol 118 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomashow MF (1999) Plant cold acclimation, freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50 571–599 [DOI] [PubMed] [Google Scholar]
- Trudel J, Asselin A (1989) Detection of chitinase activity after polyacrylamide gel electrophoresis. Anal Biochem 178 362–366 [DOI] [PubMed] [Google Scholar]
- Worrall D, Elias L, Ashford D, Smallwood M, Sidebottom C, Lillford P, Telford J, Hold C, Bowles D (1998) A carrot leucine-rich-repeat protein that inhibits ice recrystallization. Science 282 115–117 [DOI] [PubMed] [Google Scholar]
- Wu G, Robertson AJ, Liu X, Zheng P, Wilen RW, Nesbitt NT, Gusta LV (2004) A lipid transfer protein gene BG-14 is differentially regulated by abiotic stress, ABA, anisomycin, and sphingosine in bromegrass (Bromus inermis). J Plant Physiol 161 449–458 [DOI] [PubMed] [Google Scholar]
- Yeh S, Moffatt BA, Griffith M, Xiong F, Yang DS, Wiseman SB, Sarhan F, Danyluk J, Xue YQ, Hew CL, et al (2000) Chitinase genes responsive to cold encode antifreeze proteins in winter cereals. Plant Physiol 124 1251–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XM, Griffith M (2001) Winter rye antifreeze activity increases in response to cold and drought, but not abscisic acid. Physiol Plant 112 78–86 [DOI] [PubMed] [Google Scholar]
- Yu XM, Griffith M, Wiseman SB (2001) Ethylene induces antifreeze activity in winter rye leaves. Plant Physiol 126 1232–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Hrmova M, Wan CH, Wu C, Balzen J, Cai W, Wang J, Densmore LD, Fincher GB, Zhang H, et al (2004) Members of a new group of chitinase-like genes are expressed preferentially in cotton cells with secondary walls. Plant Mol Biol 54 353–372 [DOI] [PubMed] [Google Scholar]
- Zhang D-Q, Liu B, Feng D-R, He Y-M, Wang S-Q, Wang H-B, Wang J-F (2004) Significance of conserved asparagine residues in the thermal hysteresis activity of carrot antifreeze protein. Biochem J 377 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]