A phosphotriesterase (PTE) from the hyperthermophilic archaeon S. solfataricus has been crystallized. Combined with biochemical and bioengineering studies, it is expected that the structure of this protein will provide insight into the natural function of the PTE family and provide important data for achieving an efficient organophosphate biodecontaminant.
Keywords: phosphotriesterases, Sulfolobus solfataricus, organophosphate-degrading enzymes
Abstract
Organophosphates constitute the largest class of insecticides used worldwide and some of them are potent nerve agents. Consequently, organophosphate-degrading enzymes are of paramount interest as they could be used as bioscavengers and biodecontaminants. Phosphotriesterases (PTEs) are capable of hydrolyzing these toxic compounds with high efficiency. A distant and hyperthermophilic representative of the PTE family was cloned from the archeon Sulfolobus solfataricus MT4, overexpressed in Escherichia coli and crystallized; the crystals diffracted to 2.54 Å resolution. Owing to its exceptional thermostability, this PTE may be an excellent candidate for obtaining an efficient organophosphate biodecontaminant. Here, the crystallization conditions and data collection for the hyperthermophilic S. solfataricus PTE are reported.
1. Introduction
Organophosphates (OPs) are toxic compounds because they irreversibly inhibit acetylcholinesterase, a key enzyme in the nervous system. They have been distributed globally since the end of World War II and their toxic properties have been utilized in chemical warfare agents such as sarin, soman and VX and for the production of agricultural insecticides (Raushel, 2002 ▶). Enzymes that are capable of degrading OPs are currently extensively studied because of their potential in decontamination and detection systems for organophosphate-based pesticides and nerve agents (LeJeune et al., 1998 ▶). Enzymatic detoxification of OPs has become the subject of numerous studies as current methods such as bleach treatment and incineration are impractical owing to high costs or environmental concerns. Bacterial OP hydrolases are appealing for this application owing to their broad substrate specificity and high catalytic rate under mild conditions (LeJeune et al., 1998 ▶).
Enzymes that catalyse the hydrolysis of phosphoester bonds in OPs are known from several different organisms. One class of enzymes that are able to degrade nerve gases is the prolidases. As an example, a prolidase named organophosphorus acid anhydrolase (OPAA) was identified in a strain of Alteromonas (Cheng et al., 1993 ▶). Another class of enzyme, isolated from Loligo vulgaris, is diisopropylfluorophosphatase (DFPase), which prefers P—F bonds. Its three-dimensional structure was solved and reveals a six-bladed β-propeller with two calcium ions in a central water-filled tunnel (Koepke et al., 2002 ▶). HDL-associated human paraoxonase (HPON) also hydrolyses phosphotriesters, albeit with lower proficiency. The structure of HPON1 has recently been solved and shows a similar fold to that of DFPase (Harel et al., 2004 ▶). Other bacterial enzymes (EC 3.1.8.1) called phosphotriesterases (PTE) or sometimes organophosphorus hydrolases (OPH), organophosphate-degrading enzymes (OPD), parathion hydrolases (Hou et al., 1996 ▶) or paraoxonases (Merone et al., 2005 ▶), show a preference for organophosphorus compounds containing P—O or P—S bonds. Phosphotriesterases are members of the the amidohydrolase superfamily (Seibert & Raushel, 2005 ▶), enzymes that catalyze the hydrolysis of a wide range of compounds with different chemical properties (phosphoesters, esters, amides etc.). Several different structures of PTEs are available. Structures are known of the most efficient PTE, that from Pseudomonas diminuta (Munnecke, 1976 ▶), and of the very similar (90% sequence identity) OpdA from Agrobacterium radiobacter (Horne et al., 2003 ▶). Structurally, PTEs are (β/α)8-barrel enzymes with binuclear metal centres located at the C-terminal end of the barrel (Vanhooke et al., 1996 ▶). The reaction mechanism was proposed to proceed via an SN2-like mechanism in which the metal centre enables a hydroxide ion (bridged to the two metal ions) to attack the electrophilic phosphorus of the substrate (Aubert et al., 2004 ▶).
A protein from the hyperthermophilic archeon Sulfolobus solfataricus MT4, SsoPox, has recently been cloned and characterized: it is a rather distant representative of the PTE family, displaying only about 30% sequence identity to mesophilic PTEs. Despite this, all the amino acids coordinating the binuclear metal centre are conserved. It has been observed that SsoPox catalyzes the hydrolysis of paraoxon and other pesticides with a significantly lower proficiency than mesophilic PTEs, but with a similar K m. Similarly to P. diminuta PTE, its activity depends on the presence of metal cations; high activity was observed for the Co2+-substituted isoenzyme (Merone et al., 2005 ▶). SsoPox proved to have exceptional thermal stability, with denaturation half-lives of 4 h and 90 min at 368 and 373 K, respectively. This property allows high-yield purification of recombinant enzyme simply by heating cell lysates to cause precipitation of host (Escherichia coli) proteins. Recently, high catalytic activity and specificity towards lactones as substrates was reported for SsoPox and other PTE-related enzymes. Afriat et al. (2006 ▶) dubbed this new group of enzymes ‘phosphotriesterases like lactonases’ (PLLs) based on the observation of sequence features that were not present in mesophilic PTEs and of significant differences in enzyme specificity.
In particular, the activity detected towards natural homoserine lactones suggests a role in quorum-sensing signalling and led to the conclusion that PTEs evolved from a PLL-family member, probably SsoPox, utilizing its latent promiscuous phosphotriesterase activity as an essential starting point (Afriat et al., 2006 ▶). Furthermore, although its phosphotriesterase activity at ambient temperature against OPs is very low, the structure of SsoPox may provide clues to enhancing its catalytic parameters. Finally, SsoPox may be an excellent candidate in biotechnology studies seeking an efficient biodecontaminant of organophosphorus compounds.
In this report, we describe the crystallization, data collection and preliminary X-ray diffraction analysis of the hyperthermophilic S. solfataricus phosphotriesterase.
2. Crystallization
For the crystallization studies presented here, we utilized recombinant SsoPox overproduced in E. coli under the control of the T7 promoter and extensively purified in a soluble and active form (Merone et al., 2005 ▶). Crystallization assays were carried out using the hanging-drop vapour-diffusion technique (McPherson, 1990 ▶). The protein solution for crystallization was made up of 20 mM HEPES pH 8.5, 0.2 mM CoCl2 and 0.2 M NaCl. The enzyme was concentrated to 5.8 mg ml−1 using a centrifugation device (Centriprep Amicon, 10 kDa cutoff; Millipore, St Quentin-en-Yvelines, France). Equal volumes (1–2 µl) of protein and reservoir solutions were mixed and the resulting drops were equilibrated against 800 µl reservoir solution containing 15–18%(w/v) PEG 8000 and 50 mM Tris–HCl buffer pH 8. Small crystals appeared after one week at 277 K (Fig. 1 ▶). Although they did not possess clear and geometrical faces, the crystals diffracted to reasonable resolution (2.54 Å).
Figure 1.
Typical crystal of the hyperthermophilic phosphotriesterase from S. solfataricus.
3. Data collection
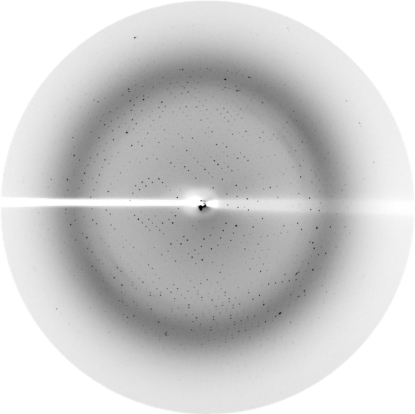
Crystals were mounted in nylon loops (Hampton Research) and flash-frozen in liquid nitrogen at 100 K using a cryoprotectant solution containing 50 mM Tris–HCl buffer pH 8, 18%(w/v) PEG 8000 and 25%(v/v) glycerol. X-ray diffraction intensities were collected at the BM30A beamline (ESRF, Grenoble, France) using a wavelength of 0.9789 Å and a MAR CCD 165 mm detector (MAR Research) with 35 s exposures. Diffraction data were collected from 180 images using the oscillation method; individual frames consisted of 1.0° oscillation steps over a range of 180° (Fig. 2 ▶).
Figure 2.
A diffraction pattern of a crystal of the phosphotriesterase from S. solfataricus. The edge of the frame is at 2.1 Å.
4. Results and conclusion
Crystals of SsoPox belong to the orthorhombic space group P212121, with unit-cell parameters a = 87.16, b = 104.82, c = 155.36 Å (Table 1 ▶). X-ray diffraction data were integrated, scaled and merged using the XDS program (Kabsch, 1993 ▶) and the CCP4 program suite (Collaborative Computational Project, Number 4, 1994 ▶). Initial molecular replacement was performed with Phaser (Read, 2001 ▶) using a polyalanine model derived from the structure of P. diminuta PTE (PDB code 1dpm; Vanhooke et al., 1996 ▶). Two protein molecules were found in the asymmetric unit (translation-function Z scores of 7.22 and 6.72). Given the molecular weight of SsoPox (35.5 kDa), this corresponds to a high Matthews coefficient (V M) of 5.0 Å3 Da−1 and a high solvent content of approximately 75.2%. The electron-density map calculated with model phases obtained from molecular replacement was of poor quality. Almost all side chains were not visible in the maps. Nevertheless, the active site constituted of two metal cations was clearly visible. The electron-density map was improved with DM (Cowtan & Zhang, 1999 ▶) using solvent flattening and noncrystallographic symmetry averaging between the two molecules (starting FOM, 0.442; FOM after DM, 0.632). Manual model improvement was performed using Coot (Emsley & Cowtan, 2004 ▶). Some side chains could be correctly placed and some loops involved in the dimer interface were removed. Using the improved model and fixing the two initial solutions, molecular replacement was performed using MOLREP (Vagin & Teplyakov, 2000 ▶) and two extra solutions emerged, giving a total of four protein molecules in the asymmetric unit (R cryst = 0.47, correlation coefficient = 0.325). This corresponds to a V M of 2.5 Å3 Da−1, which is a more typical value (Matthews, 1968 ▶). Finally, the new electron-density maps were of sufficient quality for model building. Structure refinement and interpretation are in progress.
Table 1. Diffraction data collected at BM30A FIP beamline (ESRF, Grenoble, France).
Values in parentheses are for the highest resolution shell.
| Wavelength (Å) | 0.9789 |
| Resolution (Å) | 2.54 (2.7–2.54) |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 87.16, b = 104.82, c = 155.36 |
| No. of observed reflections | 308664 (50951) |
| No. of unique reflections | 85611 (12853) |
| Completeness (%) | 95.3 (87.2) |
| Rmerge (%) | 16.0 (52.8) |
| Rmeas (%) | 11.6 (52) |
| I/σ(I) | 11.69 (3.03) |
| Redundancy | 3.60 (3.96) |
| Completeness (%) | 95.3 (87.2) |
Acknowledgments
This research was supported by grants to PM and EC from Délégation Générale pour l’Armement (CO No. 010807/03-10) and from the CNRS. We also thank MIUR project ‘Piano Nazionale Ricerca per le Biotecnologie Avanzate II–Biocatalisi’.
References
- Afriat, L., Roodvelt, C., Manco, G. & Tawfik, D. (2006). Biochemistry, 45, 13677–13686. [DOI] [PubMed] [Google Scholar]
- Aubert, S. D., Li, Y. & Raushel, F. M. (2004). Biochemistry, 43, 5707–5715. [DOI] [PubMed] [Google Scholar]
- Cheng, T. C., Harvey, S. P. & Stroup, A. N. (1993). Appl. Environ. Microbiol.59, 3138–3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Cowtan, K. D. & Zhang, K. Y. (1999). Prog. Biophys. Mol. Biol.72, 245–270. [DOI] [PubMed] [Google Scholar]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Harel, M., Aharoni, A., Gaidukov, L., Brumshtein, B., Khersonsky, O., Meged, R., Dvir, H., Ravelli, R. B., McCarthy, A., Toker, L., Silman, I., Sussman, J. L. & Tawfik, D. S. (2004). Nature Struct. Mol. Biol.11, 412–419. [DOI] [PubMed]
- Hou, X., Maser, R. L., Magenheimer, B. S. & Calvet, J. P. (1996). Gene, 168, 157–163. [DOI] [PubMed] [Google Scholar]
- Horne, I., Qiu, X., Russel, R. J. & Oakeshott, J. G. (2003). FEMS Microbiol. Lett.222, 1–8. [DOI] [PubMed] [Google Scholar]
- Kabsch, W. (1993). J. Appl. Cryst.26, 795–800. [Google Scholar]
- Koepke, J., Scharff, E. I., Lucke, C., Ruterjans, H. & Fritzsch, G. (2002). Acta Cryst. D58, 1757–1759. [DOI] [PubMed] [Google Scholar]
- LeJeune, K. E., Wild, J. R. & Russell, A. J. (1998). Nature (London), 395, 27–28. [DOI] [PubMed] [Google Scholar]
- McPherson, A. (1990). Eur. J. Biochem.189, 1–23. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Merone, L., Mandrich, L., Rossi, M. & Manco, G. (2005). Extremophiles, 9, 297–305. [DOI] [PubMed] [Google Scholar]
- Munnecke, D. M. (1976). Appl. Environ. Microbiol.32, 7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raushel, F. M. (2002). Curr. Opin. Microbiol.5, 288–295. [DOI] [PubMed] [Google Scholar]
- Read, R. J. (2001). Acta Cryst. D57, 1373–1382. [DOI] [PubMed] [Google Scholar]
- Seibert, C. M. & Raushel, F. M. (2005). Biochemistry, 44, 6383–6391. [DOI] [PubMed] [Google Scholar]
- Vagin, A. & Teplyakov, A. (2000). Acta Cryst. D56, 1622–1624. [DOI] [PubMed] [Google Scholar]
- Vanhooke, J. L, Benning, M. M., Raushel, F. M. & Holden, H. M. (1996). Biochemistry, 35, 6020–6025. [DOI] [PubMed] [Google Scholar]