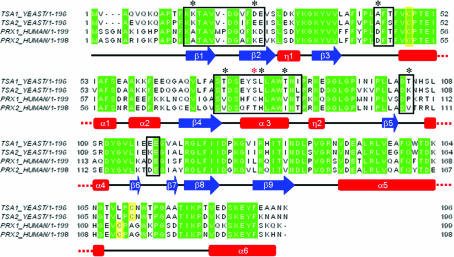
Figure 2.
Alignment of the amino-acid sequences of yeast Tsa1 and Tsa2 and the human counterparts Prx1 and Prx2. The secondary-structure assignment corresponds to the human Prx2 structure. Sequence alignment was performed using ClustalW (Thompson et al., 1994 ▶). Identical residues are highlighted in green; black boxes indicate residues involved in the dimer–dimer interface in the human Prx2 structure and their equivalents in Prx1 and yeast Tsa1 and Tsa2. Black asterisks show nonconserved residues in the dimer–dimer interface region and yellow boxes show the catalytic cysteines. The red asterisk denotes Cys83 of human Prx1.