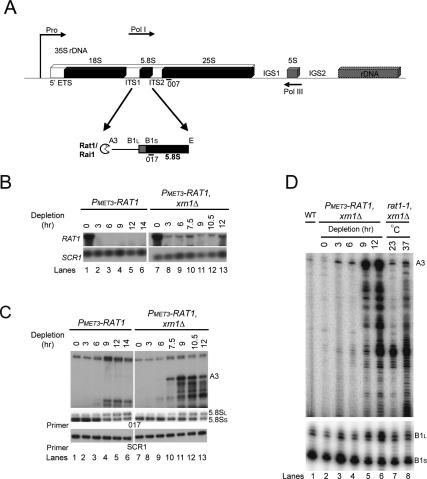
Figure 1.
Rat1 depletion leads to defects in 5.8S rRNA processing. (A) Schematic of an rDNA repeat in S. cerevisiae showing the 5′ processing of 5.8S rRNA. The rDNA repeat includes the 35S pre-RNA gene, which is transcribed by Pol I and processed to mature 18S, 5.8S, and 25S rRNAs. (Pro) Pol I promoter. The 35S genes are separated by IGS1 and IGS2 and the 5S rRNA gene, which is transcribed by Pol III in the opposite direction (indicated by arrows). The pre-rRNA is cleaved at site A3 by the endonuclease RNase MRP (Lygerou et al. 1996) and then processed by Rat1/Rai1 to site B1S, the 5′ end of the 5.8SS rRNA. A distinct minor pathway, which is independent of Rat1 activity, generates the 5′ end of the longer 5.8SL rRNA at site B1L. Processing in ITS2 generates the 3′ end of both 5.8S species at site E. (B) Rat1 mRNA depletion. Total RNA was extracted from PMET3-RAT1 and PMET3-RAT1, xrn1Δ strains grown at 30°C in the absence of methionine (0-h samples) or in the presence of 5 mM methionine for the times indicated and resolved on a 1.2% agarose/glyoxal gel. RAT1 mRNA was detected with a random primed probe (see Materials and Methods; Supplemental Table S2). (Bottom panel) As a control, the abundant cytoplasmic SCR1 RNA was detected with oligonucleotide SCR1 (see Supplemental Table S2). (C) Northern analysis of 5.8S rRNA maturation. RNAs extracted as above were resolved on an 8% polyacrylamide/urea gel. Mature 5.8SS and 5.8SL rRNAs and extended pre-5.8S species (A3) were detected with probe 017 (see Supplemental Table S2). The top panel (A3) shows a longer exposure than the middle panel (5.8SS and 5.8SL). (Bottom panel) As a control, the abundant cytoplasmic SCR1 RNA was detected with oligonucleotide SCR1. (D) Primer extension analysis of 5.8S rRNA maturation. RNA was extracted from wild-type and PMET3-RAT1, xrn1Δ strains as above and from rat1-1, xrn1Δ strain grown at 25°C and following transfer for 2 h to 37°C. Primer extension products using oligonucleotide 017 were resolved on a 6% polyacrylamide/urea gel. The primer extension stops at B1L and B1S represent the 5′ ends of the mature 5.8SS and 5.8SL rRNAs, respectively. The bottom panel shows a shorter exposure than the top panel.