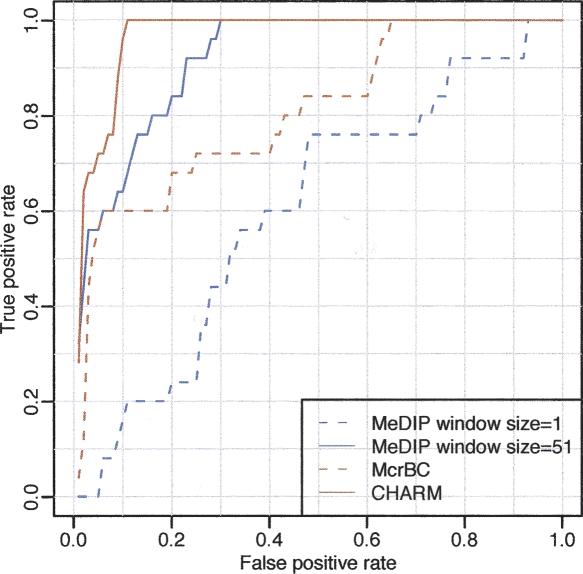
Figure 5.
ROC curve demonstrating the advantage of genome-weighted smoothing. We considered all gene regions represented on the Illumina platform. For the purpose of ROC calculation, highly methylated and unmethylated regions were compared. If all probes in the region showed on the reference Illumina platform a methylation percentage >90%, the region was considered a true positive. If all probes in the region reported a percentage <10% they were considered a true negative. To define a positive from the microarray data using a window size of 1, a cut-off for the M-values was chosen. If any probe intensity within the region was above that cut-off, it was defined as positive. A running median with a window size of 51 was then analyzed and defined a positive in the same way, except that the smoothed results instead of the individual probe intensities were used. Results are shown for both McrBC and MeDIP. For a given threshold, the true-positive rate is defined as the percentage of true-positive regions for which the microarray data surpasses that threshold. The false-positive rate is defined in the same way but for the true-negative regions.