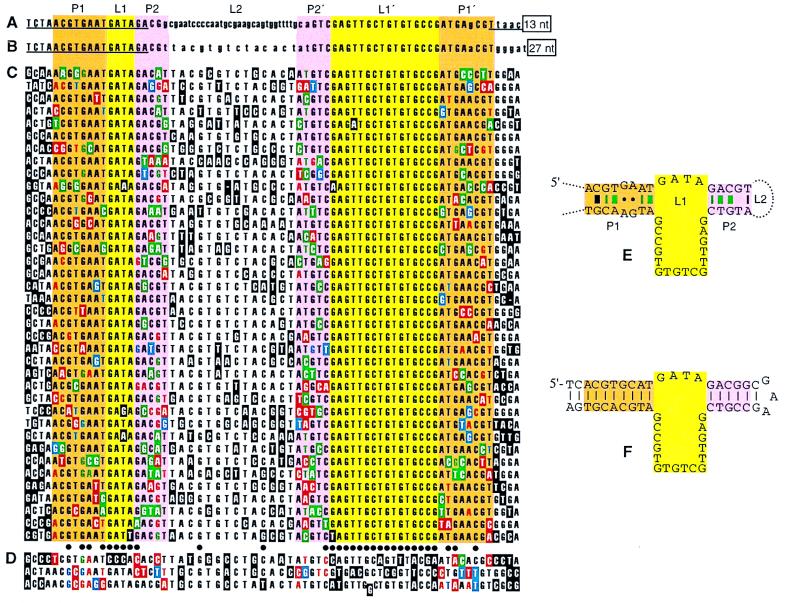
Figure 2.
A DNA motif that binds d-VP. Paired segments (orange and pink) enclose a large asymmetric internal loop (yellow), each position of which exhibits marked base preference. (A and B) The two sequences found in the final pool of the first selection experiment, aligned by large regions of sequence identity (capital letters). The central 60 positions of the initial pool were randomized; underlined sequence, serving in PCR primer binding, did not vary. A limited truncation study showed that variants lacking the boxed 3′ regions were fully active in d-VP chromatography; derivatives of sequence A lacking either 18 nt from the 5′ end or 26 nt from the 3′ end were inactive. (C and D) Aptamer variants isolated in a second selection experiment. Positions originally doped at 30% are shown aligned to the parental 68-nt segment from sequence B. Bases differing from the parental sequence are boxed. Dots mark positions where the parental base was retained in at least 35 of the 41 variants [P, the probability (binomial distribution tail) that a neutral base would be retained at this level, is 0.019]. Nearly half (43%) of the variation at these 31 conserved positions occurred in the three sequences of D. D sequences have at least nine changes from the parental base at these positions (no sequence of C has more than three such changes) and were inactive in d-VP chromatography. Coloring of boxed changes within the proposed pairing segments distinguishes whether the change would result in Watson–Crick (green), wobble (blue), or no (red) pairing; this color coding also denotes effects on pairing status of proposed partner positions that retain the parental base. It is useful to note that the ratio of changes forming new Watson–Crick pairs (green) to those disrupting pairing (red) would be 3:40 at a 30% doped Watson–Crick pair before selection. (E) Secondary structure model of sequence B. Small circles indicate putative GA:GA dinucleotide pairing. Dashes indicate where Watson–Crick or wobble pairing is maintained in ≥30 of the 38 C sequences (P = 0.011 for a parental Watson–Crick pair); dashes are colored green at very highly conserved pairs (pairing maintained in ≥33 sequences; P = 0.00031 for parental Watson–Crick pair). Thickening of dashes indicates pairs where nonparental Watson–Crick pairs occurred at significantly high levels [P ≤ 0.012 (see Materials and Methods)]. (F) Aptamer synthesized in both enantiomeric forms for subsequent study.