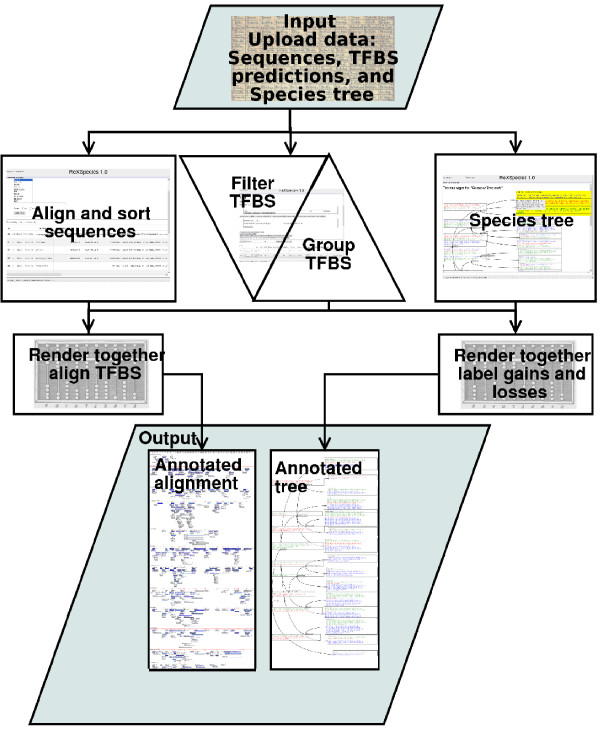
Figure 1.
Workflow overview. Uploaded sequences are processed by the alignment module. If necessary, they are aligned using Muscle [22,23] and sorted alphabetically or in a user-defined way. TFBS predictions from Mapper [1,2] and Genomatix MatInspector [3] are uploaded and then processed by the TFBS module, which places TFBS predictions onto the sequence alignment (since March 2008 TFBS predictions may be obtained directly, see "Note added in proof"). The TFBS module then filters the TFBS predictions, e.g. by E-value, by species-label of the TFBS model, or by TFBS name. Then the TFBS module groups the TFBSs by name and position. Finally the alignment, the input tree, and the TFBS predictions are put together by the rendering module resulting in an annotated alignment and an annotated species tree. Alternatively, a tree can be calculated from the alignment (including TFBS predictions) using MrBayes [18,19] and be annotated as well.