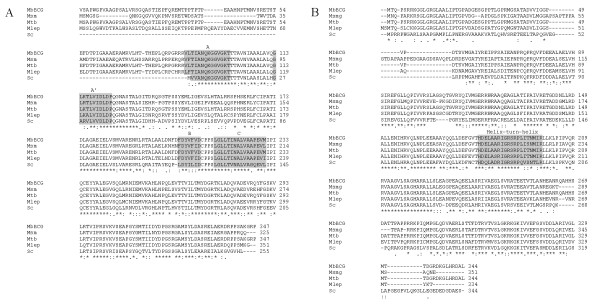
Figure 1.
Alignment of the ParA and ParB proteins. Comparison of the ParA (left) and ParB (right) aminoacid sequences of M. bovis BCG (MbBCG), M. smegmatis (Msm), M. tuberculosis (Mtb), M. leprae (Mlep) and S. coelicolor (Sc). The alignment was carried out using CLUSTAL W (1.83) http://www.ebi.ac.uk/clustalw/. Conserved amino acids are indicated with asterisk below the alignment; "-" represents gaps, ":" indicate conserved substitutions and "." semi-conserved substitutions. The A (VL/FTIANQKGGVGKT), A' (GLKTLVIDLDP) and B (FDYVFV/ID) boxes typical of the Walker-ATPases and the C motif (LGLLTINALVAAPEVM/L) of ParA proteins are highlighted on grey. The Helix-turn-Helix motif (HDELAARIGRSRPLITNMIR) involved in DNA-protein interactions of ParB is also highlighted on grey [3]. As noted, mycobacterial ParA have a longer N-terminal domain (between 67 to 91 aa) than other bacterial-ParA proteins.