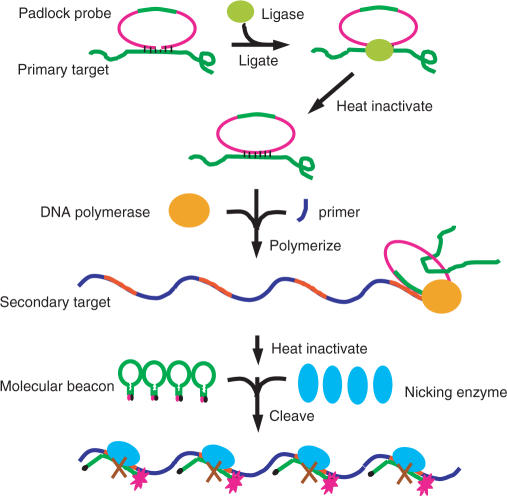
Figure 4.
Working principle of extended NESA. Basic NESA is integrated with rolling circle amplification (RCA), in order to recognize target DNA of any sequence of interest. The central element in RCA is the padlock probe, which contains, at its two ends, a target recognition sequence, and, in the middle, a sequence (green color) identical to that of the major portion (loop plus one arm) of a molecular beacon. The molecular beacon contains a nicking enzyme recognition sequence. Extended NESA includes three sequential steps: ligation, polymerization and nicking. At ligation step, a padlock probe hybridizes at two ends to target DNA and is circularized by DNA ligase. At polymerization step, a primer binds to the circularized padlock probe, and is extended by DNA polymerase, producing a long single-stranded DNA composed of tandem copies of the complementary sequence of the padlock probe, with each copy containing a complementary sequence (red color) for the molecular beacon. At nicking step, each red color sequence, like the target sequence in basic NESA, acts as a mobile catalytic site, and leads to the nicking of many molecular beacons in the presence of nicking enzymes. To be consistent with the terms used in the basic nicking assay, we designated the target DNA as the primary target, and the red color sequence in the RCA product the secondary target. In extended NESA, the primary target does not need to contain nicking enzyme recognition sequence. Extended NESA has two levels of signal amplification: each primary target induces many secondary targets through RCA, and each secondary target brings about cleavage of many beacon molecules.