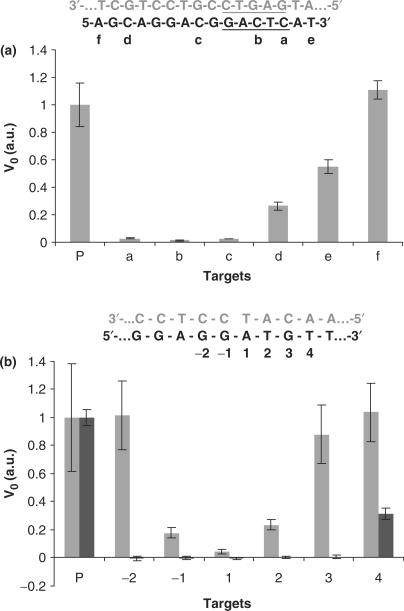
Figure 7.
Detection of single base mutation. All mutations were made by substituting the original base by its complementary base. Initial reaction rates for perfect target (P) and various mutants were compared. (a) Basic NESA. Beacon 1 was used. The sequences of beacon 1 (gray) and the target (black) are shown at the top of the chart. N.BstNB I recognition sequence is underlined. The position of mutation is labeled alphabetically. (b) Extended NESA. At the top of the chart is part of the sequence of the hybrid between the target (black) and padlock probe (gray) near the nick. Mutation positions are numbered. The nick in the padlock probe was sealed by either T4 ligase or E. coli ligase. After rolling circle polymerization, cleavage reaction was carried out, and beacon 2 was used to monitor the cleavage reaction. Two groups of results (gray and black) are presented. T4 ligase was used to obtain the gray group results, and E. coli ligase used to obtain the black group. Each datum is the average of four measurements, and the error bar is the standard deviation.