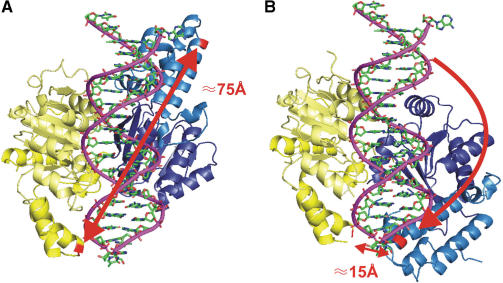
Figure 1.
Labelling sites and expected conformational changes. (A) Open conformation. A model of the SsoRad54cd based on its crystal structure is shown with a bound DNA molecule in the groove generated by the two domains (yellow and blue). Amino acid residues used for mutagenesis are shown in red. The distance between these residues is 75 Å. (B) Closed conformation. Comparison to structures of other related ATPases predicts an active conformation of the enzyme, where one domain underwent an 180° turn (10). The two labelling sites are now in very close proximity of 15 Å.