Infections due to multidrug- and carbapenem-resistant isolates of Acinetobacter baumannii were frequent among patients treated in the Tzannion General Hospital in Piraeus, Greece, throughout the period 2005 to 2006. Six of these isolates, derived from blood cultures of patients in three different wards (four were from patients in the intensive care unit) and taken at least 1 month apart from each other, were studied. Species identification, initially performed with the API 20NE system (bioMerieux, Marcy l'Etoile, France), was confirmed by sequencing of 16S rRNA genes.
Antimicrobial MICs were determined by agar dilution. Screening for metallo-β-lactamases (MBLs) was performed with imipenem-EDTA synergy tests: (i) the MBL Etest (AB Biodisk), (ii) the double-disc synergy test (3), and (iii) the combined-disc test (1). β-Lactamases were studied by isoelectric focusing of crude cell extracts derived by sonication. The carbapenemase activity of β-lactamase-containing extracts was assessed by spectrophotometry using imipenem (100 μM) as the reporter substrate and is expressed as units (1 U was the amount of enzyme hydrolyzing 1 μmol of imipenem per min per mg of protein). Reaction mixtures were supplemented with ZnSO4 (50 μM) (6). Imipenem hydrolysis rates were also determined in the presence of EDTA at a final concentration of 100 μM. Preliminary experiments using extracts from Escherichia coli laboratory strains containing various β-lactamases showed that EDTA at 100 μM inhibited VIM-1 activity by >95%, while the activities of OXA, TEM, and AmpC enzymes were virtually unaffected. The blaOXA and MBL genes were identified by PCR as described previously (10) and by sequencing. The association of blaOXA genes with ISAba elements was studied by PCR (11). blaVIM-1-carrying integrons were characterized by the sequencing of overlapping PCR products based on published sequences (7, 8). The isolates were typed by pulsed-field gel electrophoresis (PFGE) (9). The location of the VIM-1-encoding integrons was determined by an I-CeuI-based method (5).
The isolates were highly resistant to piperacillin-tazobactam, cefotaxime, ceftazidime, cefepime, and aztreonam (MICs, 64 to >128 μg/ml). Imipenem and meropenem MICs ranged from 16 to 32 μg/ml. The isolates were also resistant to ampicillin-sulbactam, gentamicin, amikacin, tobramycin, and ciprofloxacin. Only one isolate was MBL positive by the double-disc synergy test, though it tested negative by the MBL Etest and the combined-disc test. The remaining five isolates appeared to be negative by all three tests. Notwithstanding the minor phenotypic differences, the isolates exhibited similar PFGE patterns (>80% relatedness), suggesting a clonal relationship.
The isolates carried the carbapenemase genes blaOXA-51 and blaOXA-58. ISAba3 was found by PCR upstream of blaOXA-58. Moreover, ISAba1 was adjacent to blaOXA-51, probably providing the promoter for this gene (11). Other types of blaOXA genes were not identified. Unlike with the results of the MBL detection tests, all six isolates contained the blaVIM-1 MBL gene.
The results of the isoelectric focusing experiments were in line with the bla gene content. The isolates produced β-lactamases with isoelectric points (pIs) of approximately 6.5 and 7.0, consistent with OXA enzymes, as well as an enzyme with an alkaline pI (∼9.0), probably representing the chromosomal cephalosporinase of this species. A β-lactamase with a pI equal to that of VIM-1 (5.1) that was inhibited in situ by EDTA was also detected, indicating MBL expression. The levels of imipenem hydrolysis by the extracts of five blaVIM-1 carriers were similar, ranging from 28 to 36 U. The remaining blaVIM-1-positive isolate (TZ42/166) exhibited significantly higher activity against imipenem (90 U). These activities were at least 10-fold higher than those of 10 MBL-negative, carbapenem-resistant OXA-58 producers (obtained from two hospitals in Athens, Greece, in 2007) tested in parallel. EDTA reduced imipenem hydrolysis at levels similar to those of the blaVIM-1-negative isolates (1.5 to 3 U), suggesting that carbapenemase activities were largely due to VIM-1.
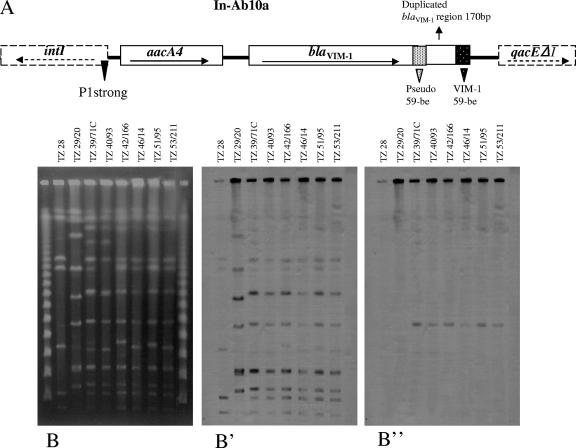
The isolates contained a class 1 integron designated In-Ab10a with a variable region of 1,725 bp comprising, from 5′ to 3′, aacA4 and blaVIM-1. The promoter region included a strong P1 and the inactive form of P2. Similar integrons have been identified in a VIM-1-encoding plasmid from an Escherichia coli isolate also from Greece (8), as well as in VIM-4-producing Pseudomonas aeruginosa isolates from Hungary and Poland (4, 7). The blaVIM cassettes in In-Ab10a and the related integrons contain, between the coding sequence and 59-be, a 170-bp direct repeat of the 3′ end of blaVIM-1. It has been proposed that this unusual structure evolved through a partial deletion of a tandem blaVIM cassette (7). In addition to In-Ab10a, isolate TZ42/166 harbored a novel integron, In-Ab10b, including only the characteristic blaVIM-1 cassette. The presence of two VIM-1-encoding integrons in TZ42/166 may explain the relatively higher carbapenemase activity; however, the higher carbapenemase activity did not seem to significantly affect β-lactam resistance levels. blaVIM-1 was detected by hybridization in an I-CeuI fragment of approximately 250 kb (Fig. 1), documenting the chromosomal location of In-Ab10a and In-Ab10b.
FIG. 1.
(A) Structure of the blaVIM-1-carrying integron In-Ab10a. The direction of transcription is indicated by the arrows. The dotted rectangle to the left of the duplicated 3′ end of blaVIM-1 (Pseudo 59-be) represents the inverse core sequence of the cassette and the pseudo-core sequence located within the structural gene. The white-dotted rectangle on the right (VIM-1 59-be) represents 59-be of the blaVIM-1 cassette. (B to B”) PFGE of I-CeuI-digested genomic DNA of eight A. baumannii clinical isolates from the same setting. (B) TZ28 and TZ29/20 in the first two lanes represent carbapenem-resistant blaVIM-1-negative isolates used for control purposes. Genomic fragments of the six blaVIM-1-positive isolates are shown in the remaining lanes. Hybridizations with probes specific for 16S rRNA and blaVIM-1 genes are shown in panels B' and B”, respectively.
The above findings indicate a protracted hospital outbreak caused by genetically related A. baumannii isolates producing VIM-1. A. baumannii isolates are routinely examined by EDTA-imipenem tests in Greek hospitals, where VIM enzymes are widespread. This approach has allowed the detection of sporadic strains carrying In-e541, a VIM-1-encoding integron that, like In-Ab10a, is also spread in enterobacteria (10). Additionally, a PCR-based screening in a hospital in northern Greece of 78 carbapenem-resistant isolates that were negative by imipenem-EDTA synergy tests revealed two blaVIM-1 carriers (2). Taken together, these studies suggest that blaVIM-1-positive A. baumannii strains have been established in this setting and that the conventional EDTA-based tests may not be suitable for their detection. The precise contribution of VIM-1 to the overall β-lactam resistance was not assessed. Nevertheless, the failure of the phenotypic tests to detect VIM-1 in these isolates underscores the role of multiple mechanisms, such as the production of OXA enzymes in determining carbapenem resistance in this species.
Nucleotide sequence accession numbers.
The sequences of the integrons In-Ab10a and In-Ab10b have been assigned GenBank accession numbers EF690695 and EF690696, respectively.
Footnotes
Published ahead of print on 19 February 2008.
REFERENCES
- 1.Franklin, C., L. Liolios, and A. Y. Peleg. 2006. Phenotypic detection of carbapenem-susceptible metallo-β-lactamase-producing gram-negative bacilli in the clinical laboratory. J. Clin. Microbiol. 44:3139-3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ikonomidis, A., E. Ntokou, A. N. Maniatis, A. Tsakris, and S. Pournaras. 21 November 2007. Hidden VIM-1 metallo-β-lactamase phenotypes among Acinetobacter baumannii clinical isolates. J. Clin. Microbiol. doi: 10.1128/JCM.01670-07. [DOI] [PMC free article] [PubMed]
- 3.Lee, K., Y. S. Lim, D. Yong, J. H. Yum, and Y. Chong. 2003. Evaluation of the Hodge test and the imipenem-EDTA double-disk synergy test for differentiating metallo-beta-lactamase-producing isolates of Pseudomonas spp. and Acinetobacter spp. J. Clin. Microbiol. 41:4623-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Libisch, B., M. Muzslay, M. Gacs, J. Minárovits, M. Knausz, J. Watine, G. Ternák, E. Kenéz, I. Kustos, L. Rókusz, K. Széles, B. Balogh, and M. Füzi. 2006. Molecular epidemiology of VIM-4 metallo-β-lactamase-producing Pseudomonas sp. isolates in Hungary. Antimicrob. Agents Chemother. 50:4220-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu, S.-L., A. Hessel, and K. E. Sanderson. 1993. Genomic mapping with I-Ceu I, an intron-encoded endonuclease specific for genes for ribosomal RNA, in Salmonella spp., Escherichia coli, and other bacteria. Proc. Natl. Acad. Sci. USA 90:6874-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loli, A., L. S. Tzouvelekis, E. Tzelepi, A. Carattoli, A. C. Vatopoulos, P. T. Tassios, and V. Miriagou. 2006. Sources of diversity of carbapenem resistance levels in Klebsiella pneumoniae carrying blaVIM-1. J. Antimicrob. Chemother. 58:669-672. [DOI] [PubMed] [Google Scholar]
- 7.Patzer, J., M. A. Toleman, L. M. Deshpande, W. Kamińska, D. Dzierzanowska, P. M. Bennett, R. N. Jones, and T. R. Walsh. 2004. Pseudomonas aeruginosa strains harbouring an unusual blaVIM-4 gene cassette isolated from hospitalized children in Poland (1998-2001). J. Antimicrob. Chemother. 53:451-456. [DOI] [PubMed] [Google Scholar]
- 8.Scoulica, E. V., I. K. Neonakis, A. I. Gikas, and Y. J. Tselentis. 2004. Spread of blaVIM-1-producing E. coli in a university hospital in Greece. Genetic analysis of the integron carrying the blaVIM-1 metallo-β-lactamase gene. Diagn. Microbiol. Infect. Dis. 48:167-172. [DOI] [PubMed] [Google Scholar]
- 9.Seifert, H., L. Dolzani, R. Bressan, T. van der Reijden, B. van Strijen, D. Stefanik, H. Heersma, and L. Dijkshoorn. 2005. Standardization and interlaboratory reproducibility assessment of pulsed-field gel electrophoresis-generated fingerprints of Acinetobacter baumannii. J. Clin. Microbiol. 43:4328-4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsakris, A., A. Ikonomidis, S. Pournaras, L. S. Tzouvelekis, D. Sofianou, N. J. Legakis, and A. N. Maniatis. 2006. VIM-1 metallo-β-lactamase in Acinetobacter baumannii. Emerg. Infect. Dis. 12:981-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turton, J. F., M. L. Ward, N. Woodford, M. E. Kaufmann, R. Pike, and T. L. Pitt. 2006. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 258:72-77. [DOI] [PubMed] [Google Scholar]