Abstract
Attachment of erythrocytes infected by Plasmodium falciparum to receptors of the microvasculature is a major contributor to the pathology and morbidity associated with malaria. Adhesion is mediated by the P. falciparum erythrocyte membrane protein 1 (PfEMP-1), which is expressed at the surface of infected erythrocytes and is linked to both antigenic variation and cytoadherence. PfEMP-1 contains multiple adhesive modules, including the Duffy binding-like domain and the cysteine-rich interdomain region (CIDR). The interaction between CIDRα and CD36 promotes stable adherence of parasitized erythrocytes to endothelial cells. Here we show that a segment within the C-terminal region of CIDRα determines CD36 binding specificity. Antibodies raised against this segment can specifically block the adhesion to CD36 of erythrocytes infected with various parasite strains. Thus, small regions of PfEMP-1 that determine binding specificity could form suitable components of an antisequestration malaria vaccine effective against different parasite strains.
The unique adhesion properties of erythrocytes infected by the Plasmodium falciparum parasite (2) play an important role in pathologies associated with severe malaria. Sequestration of parasitized erythrocytes (PEs) to specific receptors of the host vascular endothelium enables the parasite to avoid spleen-dependent clearance mechanisms. Several receptors from the host cell endothelium are involved in the mechanism of adhesion, including thrombospondin (30), intercellular adhesion molecule 1 (ICAM-1) (5), vascular adhesion molecule 1 (VCAM-1), chondroitin sulfate A (32), and CD36 (2). The combination of interactions between different receptors and the PEs contributes to the overall cytoadhesive property observed: ICAM-1 and VCAM-1 mediate an initial slowdown of circulating PEs before subsequent stable binding to receptors such as CD36 (2). Cerebral malaria, organ failure, and pregnancy-associated complications are at least in part consequences of microvascular occlusions due to the adhesion of PEs (22, 24, 39).
The antigenically diverse P. falciparum erythrocyte membrane protein 1 (PfEMP-1) (1) is encoded by the var multigene family and is expressed in a clonally variant manner at the erythrocyte surface, playing a role both in adhesion and in antigenic variation (34, 37). PfEMP-1 proteins have a mass of approximately 200 to 350 kDa and are located in knob-like protrusions at the surface of PEs, mediating the attachment of infected red blood cells to the endothelial cells of the host (1, 19). The dual role of PfEMP-1 in both sequestration and immune evasion makes it a major virulence factor of P. falciparum. A single PfEMP-1 molecule can bind to multiple receptors due to the presence of multiple adhesion domains. Various arrangements of two partially conserved binding domains, the Duffy binding-like domain (DBL) and the cysteine-rich interdomain region (CIDR), characterize each PfEMP-1 molecule (Fig. 1A), with each domain having distinct binding specificities (4, 6, 35, 36). Despite a low sequence conservation, these modules can be classified into six DBL subgroups (DBLα, DBLβ, DBLγ, DBLɛ, DBLδ, and DBLx) and three CIDR subgroups (CIDRα, CIDRβ, and CIDRγ) (16, 31). While the binding properties for some of the DBL and CIDR domains have been extensively studied, how sequence variations in the different domains precisely influence their binding specificity is still unclear (4, 6, 9, 14, 35). Major insights have recently been gained from the three-dimensional (3D) structures of two DBL domains, which recognize the Duffy antigen receptor for chemokines and glycophorin A, respectively (33, 38). In spite of a conserved overall structure, their binding properties are entirely different. No such information is currently available for any CIDR domain in PfEMP-1.
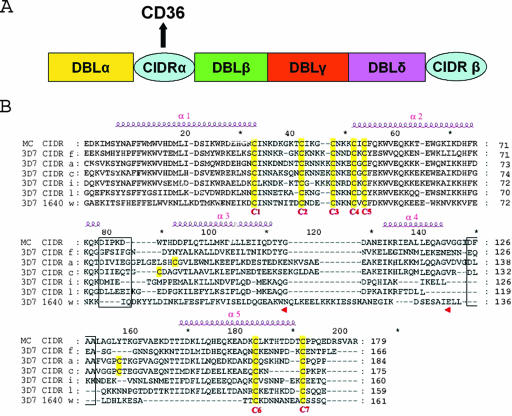
FIG. 1.
(A) Schematic representation of the modular organization in the ectodomain of a PfEMP-1 protein in the 3D7 clone of Plasmodium falciparum. The various modules of PfEMP1 that mediate cytoadhesion (DBLα, -β, -γ, and -δ and CIDRα) are indicated in different colors. The CIDRα domain mediates attachment to the CD36 receptor. (B) Amino acid sequences alignment of CIDR domains from P. falciparum strains 3D7 and MC. Accession numbers for CIDR-f and PFE1640w are given in Materials and Methods. MC CIDR (accession no. AAA89134, residues 576 to 753), CIDR-a (PFA0765c, residues 610 to 793), CIDR-c (PFD0005w, residues 633 to 807, CIDR-i (PFL0005w, residues 589 to 748), and CIDR-l (PFB1055c, residues 583 to 741) are also shown. See reference 31 for a classification of CIDR domains and their CD36 binding function. The seven conserved cysteine residues are numbered C1 to C7 (highlighted in yellow) to follow the convention introduced earlier (4). Predicted α-helices are displayed above the sequences and numbered. Putative loops that connect helices α2-α3 and α4-α5, which were deleted for binding studies (see text), are boxed.
The region within PfEMP-1 responsible for CD36 binding was initially mapped in the Malayan Camp (MC) line of P. falciparum to a 179-amino-acid fragment (MC-r179) within the central M2 region of the CIDRα domain (4) and confirmed using other parasite lines (15, 31). Comparison of various CIDR domains from several strains of P. falciparum showed that this cysteine-rich “minimal” binding domain had a conserved sequence, including the presence of five cysteine residues forming the motif CX8CX3CX3CXC (Fig. 1B). Conversely, the region recognized by CIDR molecules on CD36 maps to a hydrophobic segment located at residues 145 to 171 of the CD36 receptor (4). The importance of CD36 in the stable attachment of PEs to the endothelium suggests that disruption of this interaction would significantly interfere with the ability of the parasite to sequester. Thus, a better definition of the molecular basis of the binding specificity of CIDR domains would undoubtedly assist the development of molecules with antiadhesive properties and possibly also the development of vaccines (12, 15, 23).
The sequencing of the P. falciparum 3D7 genome provided extensive sequence information for more than 50 CIDR domains (16, 31). Based on their sequences and a solid-phase binding assay using proteins expressed at the surface of Cos-7 cells, these domains were partitioned into CIDRα domains, which can bind CD36, and non-CIDRα domains (subclassified as CIDRα1, CIDRβ, and CIDRγ), which lack this activity (31). However, the precise determinants for the binding specificity of the CIDRα domains are still elusive. We report here the cloning and expression in Escherichia coli of several CIDRα domains, including a strong CD36 binder (CIDR-f) and a previously described nonbinder (PFE1640w). Using site-directed mutagenesis and domain-swapping experiments, we demonstrated that, while the larger N-terminal part of CIDRα is essential for correct folding of the protein, a 60-amino-acid region located at the C terminus mediates binding to CD36. Interestingly, immunization of mice with this C-terminal segment of CIDRα elicits antibodies that recognize PfEMP-1 and specifically inhibit CD36 binding, for a range of different parasite strains. This work demonstrates that it is possible to generate both a cross-reactive and a potentially protective immune response using relatively small fragments of PfEMP-1.
MATERIALS AND METHODS
Sequence alignment and secondary structure prediction.
We used the MC-r179 sequence to search for similar CIDRα domains in P. falciparum clone 3D7. Fourteen amino acid sequences were aligned with MC-r179 using the program CLUSTALW, with a few manual modifications to align conserved cysteine residues. The program NNPREDICT (http://www.cmpharm.ucsf.edu/∼nomi/nnpredict.html) was used to predict the secondary structures of all CIDRα domains.
Cloning of CIDRα domains.
All PCRs were performed using genomic DNA of P. falciparum clone 3D7 as a template. The various CIDR domains were cloned into the pET-24a vector (Novagen), encoding a C-terminal hexahistidine tag, using NdeI and XhoI sites. CIDR-f (accession number PF10_0406, amino acid residues 584 to 751) and PFE1640w (accession number PFE1640w, residues 559 to 720) were amplified by PCR and cloned in frame into the vector. The full-length Duffy binding protein region II gene (accession number AAZ81536, amino acid residues 214 to 521) was subcloned from the plasmid pEGFP-HSVgD1-PvDBPII (kindly provided by John H. Adams, University of Notre Dame) into the pMAL-c2x vector (New England Biolabs) (modified through the addition of a six-histidine tag at its C terminus) between BamHI and HindIII sites. This construct was named MBP-DBPv.
Site-directed mutagenesis and domain-swapping experiment.
The mutations and primers used in this work are listed in Table 1. The various truncations are schematically represented in Fig. 2A. Domain swapping between proteins PFE1640w and CIDR-f was performed by means of a double-PCR overlapping method. Briefly, the wild-type DNA templates were purified (Qiagen) and replicated by PCR with the Turbo Pfu polymerase (Stratagene). PCR products were digested with DpnI (Stratagene) at 37°C for 2 h and transformed into SuperBlue XL-1 competent cells (Stratagene). DNA sequences for all the constructs were confirmed on an automated sequencer.
TABLE 1.
Primers used for mutagenesis studies
| CIDR f mutant (residues) | Sequences, 5′→3′ (forward, reverse) |
|---|---|
| CIDR-122Δ (1-122) | ATTTACATATGAAGGAAGAAAAAAGTATGC, ATTCTCGAGTTCATTTTCCTTTTCC |
| CIDR-105Δ (1-105) | ATTTACATATGAAGGAAGAAAAAAGTATGC, ATTCTCGAGTCCATAAGTATCTTTAATATTTG |
| CIDR-Δ106 (106-166) | ATTACATATGAATGTAAAGGAATTAGAAG, AATCTCGAGAGGGGTATTTTCGCAT |
| CIDR-Δ122 (122-166) | ATTACATATGAATGTAAAGGAATTAGAAG, ATTCTCGAGTCCATAAGTATCTTTAATATTTG |
| 1640-f chimera | CTAGCTAGCGGCGCGGAAAAAATA, CTAATTCCTTTACATTTTTAGAAGACTCAATT; AATTGAGTCTTCTAAAAATGTAAAGGAATTAG, AATCTCGAGAGGGGTATTTTCGCAT |
| f-1640 chimera | ATTACATATGAATGTAAAGGAATTAGAAG, GCACTTTCTGAATCTTTAATTCCTTCATTTC; GAAAGACTTAGAAATTAAGGAAGTAAAC, TATCTCGAGCTGGGAAGAGGAACATGC |
| DeleGFSIFG | TAAAAAACAAGGAAATGATTATAATTATGCTCTTAAAGC, GCATAATTATAATCATTTCCTTGTTTTTTAAAATGCTG |
| Dele EQEA | GGAAAATGAAAATAATTCTGGTGGCAATAACAGTC, GTTATTGCCACCAGAATTATTTTCATTTTCCTTTTCC |
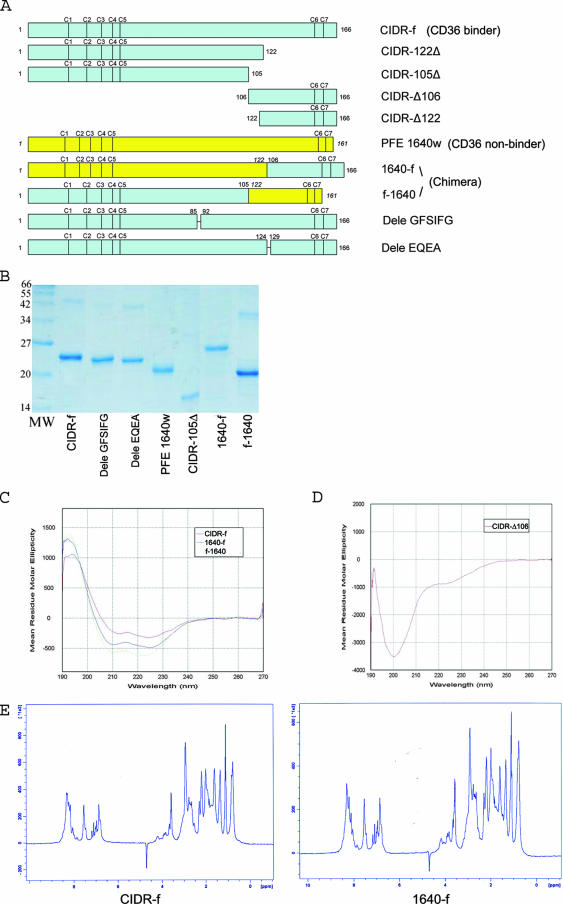
FIG. 2.
(A) Schematic view of the various CIDR constructs and chimeric proteins used for the binding experiments. CIDR-Δ106 and CIDR-Δ122 were expressed in E. coli both as GST fusion proteins and as C-terminal His-tagged fragments (see text). The positions of conserved cysteine residues are indicated. (B) Expression and purification of the various CIDRα mutants and chimeric proteins. All proteins were purified by Ni-nitrilotriacetic acid affinity, ion-exchange, and size-exclusion chromatography and separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. (C) CD spectra of key CIDR constructs and chimeric proteins. The CIDR-f protein and the two chimeric proteins 1640-f and f-1640 all showed similar spectra with one maximum at 190 nm and two minima at 208 nm and 222 nm, indicative of predominantly α-helical proteins. (D) By contrast, the His-tagged CIDR-Δ106 fragment is devoid of secondary structures. (E) 1D 1H NMR spectra of CIDR-f and 1640-f (the CD36 binding active chimera [see text]). The NMR spectra of these two recombinant proteins showed a clear dispersion of resonance lines in the region of amide protons (between 6 and 10 ppm) but downfield-shifted methyl protons (−0.5 to 1.5 ppm) resonances, indicative of globular folded proteins.
Protein expression and purification.
CIDR-f, as well as all mutants and truncated constructs, was transformed into E. coli Rosetta-gami (DE3) cells (Novagen) and subjected to the same purification protocol. Protein expression was induced through the addition of a final concentration of 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG), followed by overnight incubation at 20°C before harvesting the cells. Cells were disrupted by sonication and purified under native conditions using Ni-nitrilotriacetic acid resin (Qiagen), followed by ion-exchange chromatography using a Hiprep QFF column (Amersham) and a Superdex 200 GL size exclusion chromatography column (Amersham). A recombinant soluble human CD36, encompassing residues Gly-30 to Asn-439 (fused with a human immunoglobulin G1 Fc fragment at its C terminus and hereafter called CD36/Fc) was purchased from R&D Systems and used for all binding experiments.
CD.
Circular dichroism (CD) experiments were carried out using a ChiraScan CD spectrophotometer (Applied Photophysics) with a 1-mm cell at room temperature. Spectra were collected using wavelengths ranging from 190 nm to 260 nm with steps of 0.1 nm. All proteins were dissolved in phosphate-buffered saline at pH 7.4 to a final concentration of 0.5 mg/ml. CD spectra were corrected by subtracting the spectrum obtained from the buffer solution.
1D 1H NMR spectroscopy.
For all constructs subjected to 1D 1H nuclear magnetic resonance (NMR) spectroscopy, a 500-μl solution containing 200 μM of protein was prepared in an aqueous buffer containing sodium phosphate buffer, 50 mM NaCl, 1 mM dithiothreitol at pH 6.0, and 10% D2O (vol/vol). The NMR spectra were collected at 285 K on an Avance600 spectrometer (Bruker, Billerica, MA). The spectra were processed and analyzed with the program ZGGPW5 (Bruker).
Enzyme-linked immunosorbent assay (ELISA)-based CD36 binding assay.
Human CD36/Fc protein was applied at a concentration of 0.2 μg/ml to a MaxiSorp Immuno 96-well plate (Nunc) overnight at 4°C. After 2 hours of blocking, a range of concentrations of purified CIDRα proteins or mutants thereof were incubated for 1 h at 37°C. The amount of protein bound to the immobilized CD36/Fc protein adsorbed on the well was then revealed using a Penta-His antibody (Qiagen), followed by an alkaline phosphatase-conjugated goat anti-mouse immunoglobulin G (Sigma). Finally, Immuno Pure (p-nitrophenyl phosphate disodium salt) tablets (Pierce) dissolved in diethanolamine buffer were used for signal detection with a microplate spectrophotometer (Bio-Rad).
Peptide inhibition assay.
For the peptide inhibition assay, two overlapping peptides, CD36:145-171 (ASHIYQNQFVQMILNSLINKSKSSMFQ) and CD36:146-164 (SHIYQNQFVQMILNSLINK), derived from the CD36 sequence (3), and an unrelated peptide (KAFTTTLRGAQRLAALGDTA) were synthesized by the 9-fluorenylmethoxy carbonyl method using an automatic peptide synthesizer (Intavis AG, Bioanalytical Instruments) and purified by high-performance liquid chromatography (Shimadzu) using a C8 column (Agilent). The peptide sequences were confirmed by mass spectrometry. After preincubation of CIDR-f, PFE1640w, 1640-f, or f-1640 chimera at room temperature for 1 h at a constant concentration of 2 μg/ml with peptide concentrations ranging from 10 nM to 1 mM, the mixtures were added to a 96-well plate that had been previously coated with CD36/Fc and incubated for 2 h at 25°C. Penta-His antibody (Qiagen) and alkaline phosphatase-conjugated goat anti-mouse antibody were used to determine inhibitory effects of peptides on CIDRα and CD36/Fc binding.
Generation of polyclonal antibodies against the C-terminal end of CIDR-f.
For raising antibodies against the C-terminal end of CIDR-f, two constructs, GST-CIDR-Δ106 and GST-CIDR-Δ122 were made, both having glutathione S-transferase (GST) at their N termini. The GST-CIDR-Δ122 protein has a deletion corresponding to the region NVKELE between helices α3 and α4. Both of these two fusion proteins were expressed in E. coli and purified using glutathione-Sepharose (Amersham) according to the manufacture's protocol. Purified GST fusion proteins (50 μg per mouse) were injected into male 7- to 8-week-old BALB/c mice with complete Freund's adjuvant (Pierce) for the first injection and with incomplete Freund's adjuvant for the subsequent injections at 4-week intervals. Blood was drawn from the animals 2 weeks after each boost and tested for antibody titer by ELISAs on 96-well polystyrene plates and Western blotting.
Antiserum inhibition binding assay.
A procedure similar to the peptide inhibition assay was used for evaluating the inhibition effect on CD36/Fc and CIDR-f binding by the antisera. Human CD36/Fc proteins were applied at a concentration of 0.2 μg/ml (0.02 μM) to a MaxiSorp Immuno 96-well plate (Nunc) overnight at 4°C. After preincubation of a constant concentration of CIDR-f (4 μg/ml) with serial dilution of two antisera from 1:8 to 1:1,024 at room temperature for 2 h, the mixtures were added to a 96-well plate and incubated for 2 h at 37°C. The amount of CIDR-f binding to the plate was detected using a Penta-His horseradish peroxidase conjugate (Qiagen), followed by the horseradish peroxidase substrate o-phenylenediamine dihydrochloride (Sigma), and the optical density at 492 nm (OD492) was read. Preimmune serum, anti-GST sera (Amersham), and an irrelevant serum (a rhoptry protein polyclonal antiserum) were used as controls.
Mammalian cell culture and parasite culture.
Human lung endothelial cells (HLECs) and Chinese hamster ovary K1 cells (CHO-K1) were cultured using RPMI 1640 medium with 10% fetal bovine serum. P. falciparum clones 3D7, HB3, and FCR3 were grown in human erythrocytes from malaria-negative donors with daily changed RPMI medium under standard conditions as described previously (28), replacing 10% human serum with 2.5% Albumax. Highly synchronized parasites in mature blood-stage-infected erythrocytes of the CD36 or CSA adhesive phenotype were obtained by regular panning on HLECs or CHO-K1 cells as described elsewhere (28).
Western blotting of parasite extracts.
Parasites were cultivated to late trophozoite-schizont stages and extracted as described previously (11). The extracted mature-stage parasites were lysed directly into sample buffer and frozen-thawed three times. The supernatants were separated using a 4% to 12% NuPAGE gradient gel (Invitrogen). After the proteins were transferred onto nitrocellulose membranes, polyclonal antisera against CIDR-Δ106 fused to GST were used to detect the expression of PfEMP-1 at a dilution of 1:200, followed by secondary antibodies and enhanced chemiluminescence (Pierce).
Liquid-phase immunofluorescence microscopy.
After three washes of mature blood-stage-infected erythrocytes with culture medium without Albumax, the PEs were incubated with polyclonal antiserum against GST-CIDR-Δ106 at a dilution of 1:20 to 1:50 at 4°C for 30 min. The PEs were then incubated at 4°C for an additional 30 min with a fluorescein isothiocyanate-conjugated goat anti-mouse antibody (Jackson Immunoresearch Laboratory) after three washes with phosphate-buffered saline. Preimmune sera and the anti-GST antibody were used as negative controls. Vectorshield mounting medium with 4,6-diamidino-2-phenyl-indole dihydrochloride (DAPI) (Vector Laboratories) was applied to the slides. Immunofluorescence staining was analyzed with an Olympus fluorescence microscope.
PE adhesion inhibition assay using recombinant proteins and polyclonal antisera.
HLECs and CHO-K1 cells were cultured in 48-well plates for at least 3 days at approximately 80% confluence. Late trophozoite-stage PEs diluted in binding buffer (RPMI 1640, HEPES, 10% fetal bovine serum, pH 6.8) at a concentration of 5 × 106/ml were incubated with different dilutions of polyclonal antisera against GST-CIDR-Δ106 for 30 to 45 min at room temperature before being added to HLEC/CHO-K1 cells. HLECs/CHO-K1 cells were incubated with different recombinant proteins at concentrations ranging from 1.5 μM to 12 μM for 30 min at room temperature before parasite addition. After 60 min of incubation interspersed with gentle resuspension at 15-min intervals, the plate was washed and fixed using 2% glutaraldehyde. The average number of PEs bound to 100 HLECs or CHO-K1 cells for three microscopic fields (magnification, ×40) was calculated, and the percent inhibition of parasite binding compared to the untreated control was determined. Each assay was performed in triplicate.
RESULTS
Expression and characterization of CIDR domains.
Previous work led to a partition of CIDR domains into CD36 binders and nonbinders and identified a minimal region of CIDR required for specific binding to CD36 (4, 31). To obtain further insight into the molecular basis of the different binding phenotypes, we selected six different CIDR domains of P. falciparum clone 3D7 representing five CD36 binders and one nonbinder (Fig. 1). We aligned their sequences with the previously characterized minimal binding domain MC-r179. All sequences contain seven strictly conserved cysteine residues (sequentially numbered from 1 to 7 to match the nomenclature proposed earlier [4]) (Fig. 1B). The amino acid sequence identity the between CD36 binders and the nonbinder is ∼23 to 31%. Similar values (∼28 to 42%) are found when comparing CD36 binders. Despite their relatively low sequence identities, secondary structure predictions suggest a conserved, mainly α-helical structure for the CIDRα domains studied here, with four to five α-helices (Fig. 1B). The CIDR-f protein and the nonbinder PFE1640w (Fig. 1B) were expressed and purified in soluble form from E. coli (Fig. 2B) and analyzed by CD (Fig. 2C and data not shown). CD spectra showed a similar pattern with a maximum at 190 nm and two shallow minima at 208 nm and 222 nm, regardless of their capacity to bind CD36. These spectra are typical of proteins with a high content of α-helices, a feature consistent with secondary structure predictions. To further check that the different recombinant CIDRα domains were properly folded, we used 1D 1H NMR spectroscopy (27). The results showed a clear dispersion of resonance lines in the region of amide protons (between 6 and 10 ppm) but downfield-shifted methyl proton (−0.5 to 1.5 ppm) resonances, indicating proper folding for the CIDR-f recombinant protein (Fig. 2E, left panel). However the spectra (Fig. 2E) also confirmed a tendency of CIDR domains to oligomerize or aggregate, as shown by a comparison with typical 1D NMR spectra obtained from a large group of proteins subjected to crystallization trials (27). To evaluate the ability of the different recombinant CIDR proteins to bind to CD36, we used the ELISA method, which allows semiquantitative studies on protein-protein interactions (18). Of the six different CIDR regions that were expressed well, CIDR-f (Fig. 3A) as well as CIDR-c and CIDR-i (data not shown) bound CD36 in a concentration-dependent manner, while the CD36 nonbinder PFE1640w along with the P. vivax Duffy binding protein (MBP-DBPv) (negative control) did not (Fig. 3A). This confirmed the predicted binding properties of the expressed CIDR domains. Due to extensive degradation, CIDR-a and CIDR-l could not be used in this binding experiment. Since we observed no difference in the binding to CD36 among the various CIDR domains tested, all subsequent studies focused on CIDR-f.
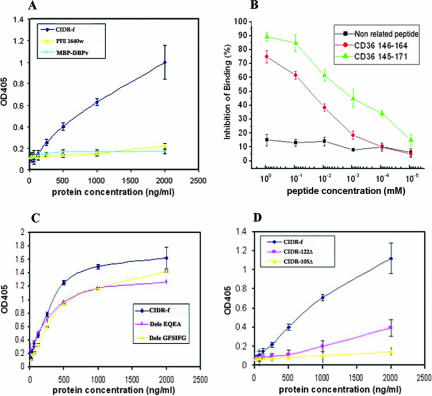
FIG. 3.
(A) Direct binding of CIDR-f protein to CD36/Fc. Neither the PFE1640w protein nor the DBP (negative control) show significant binding to CD36, with which the well was coated. Results are expressed as means and standard deviations from three independent experiments. (B) Peptide inhibition assay of CIDRα-CD36 binding. Three peptides, i.e., CD36:145-171, CD36:146-164, and a nonrelated peptide, were tested for their effects on inhibition of the interaction between CIDR-f and CD36/Fc. The percentage of binding inhibition was calculated as the ratio of OD405 in the presence of competitor to OD405 of the control sample (in the absence of peptide competitor). (C) Ability of loop deletion constructs to bind to CD36. (D) Binding of C-terminally truncated CIDR-f domains to CD36.
In order to confirm the specificity of the interaction between the recombinant CIDR-f domain and the CD36 receptor, competition assays using the previously identified region of the human CD36 receptor were performed. Two overlapping synthetic peptides, CD36:145-171 and CD36:146-164, derived from the CD36 ectodomain sequence (3) effectively disrupted the binding between CIDR-f and CD36 in a concentration-dependent manner, while an unrelated peptide did not (Fig. 3B). Peptide CD36:145-171 showed about 85% inhibition at the concentration of 100 μM, while CD36:146-164 only showed about 60% inhibition at the same concentration. The difference is probably caused by the poor solubility of peptide CD36:146-164. A 50% inhibition of binding was observed at a concentration of ∼1 μM of CD36:145-171 peptide.
The C-terminal region of CIDRα is the main determinant for CD36 binding.
According to the sequence analysis (Fig. 1B), two putative loops connecting helices α2 and α3 (86-GFSIFG) and α4 to α5 (125-EQEA) vary significantly in length and amino acids. To assess their implication in binding, we designed CIDR constructs devoid of these two putative loops (Dele GFSIFG and Dele EQEA). We also designed constructs devoid of the putative C-terminal α-helix α5 (construct CIDR-122Δ) or containing neither helix α4 nor α5 (construct CIDR-105Δ) (Fig. 1B and 2A). These proteins were expressed in a soluble form and purified (Fig. 2B and data not shown). Their CD spectra indicated the preservation of a predominantly α-helical structure (data not shown). In the ELISA-based binding assay, deletion of either loop reduced binding activity by approximately 25 to 35% compared to the full-length protein, suggesting a limited involvement of these putative loops in binding (Fig. 3C). By contrast, deletion of helix α5 led to a reduction of binding by 60 to 70%, and the additional truncation of putative helix α4 completely abolished the interaction with CD36 (Fig. 3D). The lack of CD36 binding activity of CIDR-105Δ was not due to a collapse of the whole structure, as the CD spectra still showed a predominant helical structure, comparable to that of the wild-type protein (data not shown).
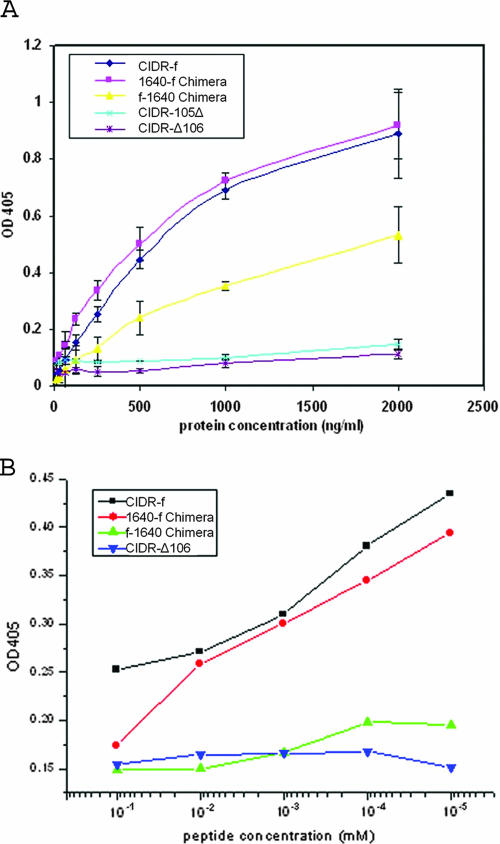
From this analysis, we conclude that the region between residues 106 and 166 of the CIDRα molecule plays a major role in CD36 binding. To establish whether this region alone could interact with CD36, we expressed the C-terminal fragment of CIDR-f as a His-tagged protein spanning residues 106 to 166 (CIDR-Δ106) (Fig. 2A) and assayed it for CD36 binding. No binding could be detected (Fig. 4A). Consistently, its CD spectra indicated that this fragment is unfolded (Fig. 2D). This suggests that while the C-terminal region is an important determinant of binding specificity, other parts of the CIDR molecule may play a role in its folding into a functional CD36 binding domain. In order to demonstrate that this region contains the structural determinants necessary and sufficient for strong binding to the CD36 receptor, we constructed two chimeric CIDR molecules (Fig. 2A): one fused the N-terminal end sequence (residues 1 to 122) of the non-CD36 binder PFE1640w (15, 31) with the C-terminal region (residues 106 to 166) of the CIDR-f molecule (1640-f chimera), while the other fused the N-terminal region (residues 1 to 105) of CIDR-f with the C-terminal region (residues 122 to 161) of the nonbinder PFE1640w (f-1640 chimera) (Fig. 2A). These two chimeric CIDR molecules were expressed well (Fig. 2B), and their CD spectra showed a content of secondary structure elements comparable to that of the wild-type CIDR-f protein (Fig. 2C). Remarkably, 1640-f converted the non-CD36 binder to about 98% of wild-type levels of CD36 binding activity, while the f-1640 chimera showed only minimal binding activity (Fig. 4A). To further investigate whether 1640-f is specific for the same region of the CD36 molecule that is bound by CIDR-f, we tested the ability of the CD36 peptide from residue 145 to 171 to compete for the interaction. As shown in Fig. 4B, this peptide is able to compete equally well for CD36 binding with either CIDR-f or the 1640-f chimera. By contrast, a minimal or no effect is seen for either the f-1640 chimera or the CIDR-Δ106 fragment, confirming the specificity of 1640-f for CD36 (Fig. 4B).
FIG. 4.
(A) CD36 binding assays for different CIDR chimeric constructs. CIDR-105Δ and CIDR-Δ106 totally lost binding ability (see Fig. 2A for the nomenclature of the constructs). The 1640-f chimera binds as well as CIDR-f to the CD36 receptor molecule. Results are expressed as means and standard deviations from three independent experiments. (B) Peptide inhibition of the interaction between CIDR and CD36. The binding of both CIDR-f and the 1640-f chimera to CD36 is competed by the CD36:145-171 peptide in a concentration-dependent manner.
CIDR domains specifically inhibit CD36 but not CSA binding of different clones of P. falciparum.
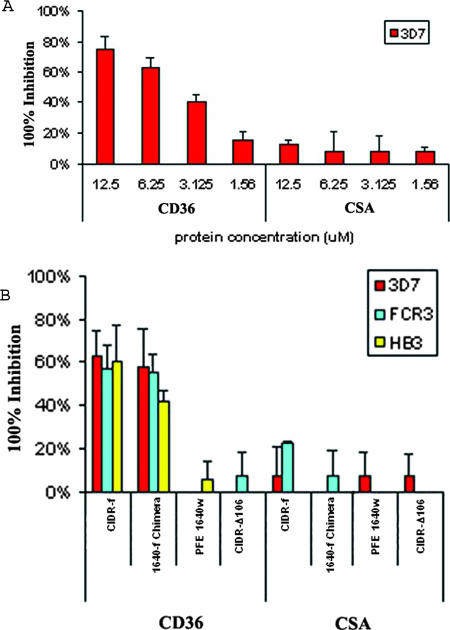
To establish whether the ELISA-based in vitro binding assays did reflect differences in the abilities of the different CIDR constructs to bind to CD36 under in vivo-like conditions, we utilized HLEC and CHO-K1 cell lines that express either CD36 or CSA on their surface. P. falciparum-infected erythrocytes were selected for a number of cycles on either cell line to enrich for parasite populations that predominately expressed PfEMP-1 molecules binding either CD36 or CSA (20, 28). The ability of the different recombinant CIDR constructs described above to inhibit the attachment of these selected parasites was then determined. Preincubation of the HLEC-CD36 cell line with CIDR-f inhibited binding of the CD36-selected 3D7 parasites in a concentration-dependent manner, while preincubation of the CHO-K1 cell line with the same protein followed by the addition of CSA-selected parasites showed no inhibition (Fig. 5A). Fifty percent inhibition of binding to CD36 was seen at a concentration of 5 to 6 μM of CIDR-f, a result in line with previous studies (10, 40). We also tested inhibition of PE adherence of P. falciparum strains 3D7, HB3, and FCR3, selected for either CD36 or CSA binding, using truncated and chimeric proteins at a concentration of 6 μM. At this concentration CIDR-f as well as 1640-f showed 40 to 60% inhibition of PE adherence to HLEC-CD36 in all three strains, while all the other recombinant proteins tested showed negligible inhibition (Fig. 5B). None of the proteins tested showed any significant inhibitory effects (0 to 20%) on CSA-adherent PEs of three strains (Fig. 5B). These results are consistent with our ELISA-based binding assay, confirming that the CD36 binding specificity is located in the C-terminal region of CIDR-f.
FIG. 5.
(A) Blockade of CD36-adherent PEs of strain 3D7 using recombinant CIDR-f proteins. Recombinant CIDR-f proteins at 1.56 μM to 12 μM were preincubated with HLECs or CHO-K1 cells in the binding buffer. After 30 min of incubation, both CD36-adherent and CSA-adherent PEs of strain 3D7 were added to each well and tested for binding. The average number of PEs bound to 100 HLECs or CHO-K1 cells in three microscopic fields (magnification, ×40) was calculated. The percentage of inhibition of parasite binding was compared to that for the untreated control without the addition of any proteins. (B) Blockade of CD36-adherent PEs of strains 3D7, HB3, and FCR3 using recombinant proteins CIDR-f, 1640-f chimera, PFE1640w, and CIDR-Δ106. A quantity of 6 μM of each recombinant protein was preincubated with HLECs or CHO-K1 cells in the binding buffer. The average number of PEs bound to 100 HLECs or CHO-K1 cells in three microscopic fields (magnification, ×40) was calculated. Standard deviations were determined from three independent experiments.
Antibodies raised against GST-CIDR-Δ106 inhibit CD36-CIDRα binding for a number of different parasite strains.
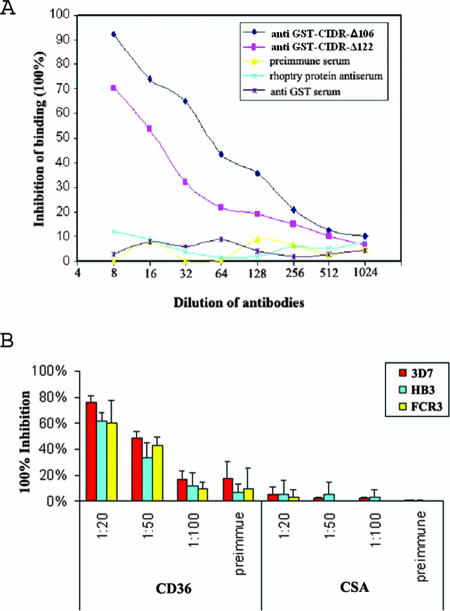
Previous work and the study presented here clearly demonstrate that recombinant PfEMP-1 CIDR can prevent a variety of strains of P. falciparum-infected erythrocytes from binding CD36 (10). This is in stark contrast to the properties of antibodies raised against the same recombinant protein, where inhibition of CD36 binding is observed to be predominantly strain specific (4). Previous work focused on immunizations using functional full-length CIDR domains. We investigated here whether smaller fragments of CIDR could circumvent this strain specificity observed in the antibody response. For this purpose, BALB/C mice were immunized with either GST-CIDR-Δ106 or GST-CIDR-Δ122. Antibodies against both proteins gave similar titers when assessed by ELISA and specifically reacted with the corresponding proteins by Western blotting (data not shown). Both antisera significantly inhibited CIDR-f binding to CD36, in a concentration-dependent manner, with the antiserum raised against GST-CIDR-Δ106 giving a higher level of inhibition than the antibodies raised against GST-CIDR-Δ122 (Fig. 6A). No inhibition of binding was observed with either a preimmune serum, anti-GST antibodies, or an irrelevant antibody directed against a parasite rhoptry protein (Fig. 6A). As the antiserum raised against GST-CIDR-Δ122 was less efficient than the one raised against GST-CIDR-Δ106, subsequent studies were performed using the latter.
FIG. 6.
(A) Antiserum inhibition assay. Sera raised against GST fused with CIDR-Δ106 and GST fused with CIDR-Δ122 were tested for inhibition effects on CIDR-f and CD36/Fc binding. The percentage of binding inhibition was calculated as the ratio of the OD405 of a control sample (without any serum) to the OD405 of each sample with various serum dilutions. (B) Blockade of CD36-adherent PEs of strains 3D7, HB3, and FCR3 using murine polyclonal antisera against CIDR-Δ106. PEs were preincubated with binding buffer alone or binding buffer containing antisera diluted from 1:20 to 1:100 or preimmune sera. After 30 of min incubation, the PEs were added to HLECs or CHO-K1 cells and tested for binding. The average number of PEs bound to 100 HLECs or CHO-K1 cells in three microscopic fields (magnification, ×40) was calculated. The percent inhibition of parasite binding compared to that for the untreated control without any added proteins was determined. Standard deviations were determined from three independent experiments.
To establish whether the antisera were also able to inhibit binding of PEs to CD36 and whether they had cross-reactive effects toward different strains of P. falciparum, we investigated their effect on the interaction between CD36-adherent or CSA-adherent PEs and HLEC or CHO-K1 cells. Antisera against GST-CIDR-Δ106 inhibited more than 60% of the binding between mammalian-expressed CD36 and CD36-adherent PEs at a dilution of 1:20 in strains 3D7, HB3, and FCR3, while at the same time showing no significant inhibitory effects on CSA binding PEs, indicating both specificity as well as cross-reactivity of this antiserum (Fig. 6B).
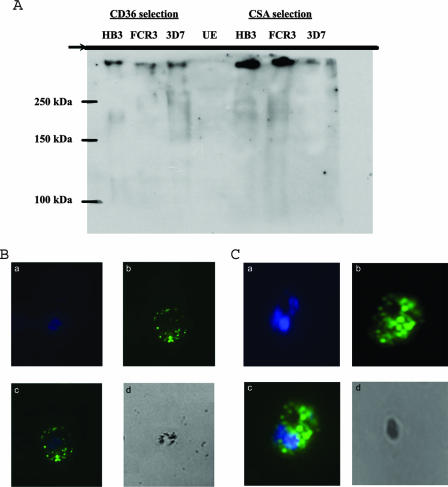
The polyclonal antibodies could recognize full-length PfEMP-1 from these different parasite strains: a specific protein of >250 kDa was detected by the anti-GST-CIDR-Δ106 sera in all the protein extracts from different parasite strains that had been selected for either CD36 or CSA (Fig. 7A). Some small size variations between the protein detected in the 3D7 strain selected on CD36 and that selected on CSA might be related to the expected change in PfEMP-1 expression (31, 36). Immunofluorescence microscopy using the same sera on CD36-adherent and CSA-adherent PEs from strain 3D7 gave a staining pattern consistent with PfEMP-1 expression (Fig. 7B and C) (14, 20), with the corresponding preimmune or anti-GST sera showing no fluorescence (data not shown). Taken together, these data showed that the sera recognized CIDR domains of PfEMP-1 from different parasite strains.
FIG. 7.
(A) Western blot of PEs using the anti-GST-CIDR-Δ106 polyclonal antibodies. Mature-stage parasites were extracted and lysed. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes. The top of the separating gel is indicated. Murine polyclonal antisera against GST-CIDR-Δ106 were used to detect the expression of PfEMP-1 at a dilution of 1:200, followed by secondary antibodies and enhanced chemiluminescence. PfEMP-1 can be detected on both CD36-adherent and CSA-adherent PEs of strains 3D7, HB3, and FCR3, while no signal can be detected in uninfected erythrocytes. (B and C) Surface labeling of CD36- and CSA-adherent PEs of strain 3D7 with murine polyclonal antisera raised against GST-CIDR-f:106-166. a, staining of parasite nuclei using DAPI; b, surface labeling of fluorescein isothiocyanate; c, merged image; d, phase image.
DISCUSSION
Antigenic variation and other immune escape mechanisms as well as our limited understanding of parasite-host interactions have made the development of an efficient malaria vaccine an exceptionally challenging task. Antibodies to PfEMP-1 correlate with the development of clinical immunity (7, 8, 17, 29). Among the various modules present in PfEMP-1, CIDR domains are attractive target for vaccine development. Since CD36 is a critical receptor, disruption of the interactions between CIDRα and CD36 with small-molecule compounds or by specific antibodies that would bind to either partner holds great promise for the development of new therapeutic strategies against malaria. To date, there are no experimental data that precisely map the region of the CIDRα domain which contributes to CD36 binding. Attempts have been made to delineate residues in the CIDRα domain of the MC clone, which is involved in CD36 binding (15, 31). The single mutations Q648K, EQ727LK, S596T, R600N, K611T, K629R, and K640S appear not to greatly affect CD36 binding, but a combination of mutations as well as a 3-amino-acid change from DIE to GHR in parasite strain ItG2-CS2 reduced the CD36 binding capacity to 63% of the wild-type level and abrogated the total binding capacity (15, 31). Although these studies could not rule out an incorrect folding of the mutant proteins, they strongly suggest that several discontinuous elements of the CIDR domain are brought together to form a topographical binding site for CD36. Our data provide strong evidence that the C-terminal region of CIDRα is the sole determinant for the CD36 binding specificity. This is reminiscent of immunoglobulin molecules, where a limited number of loops variable in length and in sequence determining the binding specificity of the molecule are presented on an essentially conserved structural framework (21, 26). Work using the MC-r179 recombinant protein demonstrated that the binding of PEs from a variety of parasite lines to CD36 can be specifically inhibited by competition with the recombinant protein and that the addition of the protein to an in vivo mouse model led to the desequestration of PEs (40). This contrasted with the inhibitory effect of antibodies raised against the same protein, where adherence of PEs was inhibited only in a strain-specific fashion, with limited effects seen on CD36 binding of PEs other than the MC strain (4). This is consistent with the role of PfEMP-1 in antigenic variation, with antibodies directed against a variant from one strain having limited cross-reactivity against another. Antibodies raised against the full-length MC r-179 protein had only a strain-specific inhibitory reaction (4), while antibodies against short peptides within MC r-179 induced cross-reactive inhibition effects. Monoclonal antibodies raised against residues 1 to 87 and 81 to 141 of MC r179 reacted with multiple P. falciparum strains expressing variant PfEMP-1 (13). Furthermore, immunization with recombinant CIDRα could protect Aotus monkeys against severe disease (23), while antibodies raised against a recombinant DBL 1α domain did generate some cross-protective antibodies (25).
While these approaches have given credence to the idea that vaccine development focusing on regions of the variant PfEMP-1 is indeed feasible, they suffered from the inability to produce truly cross-reactive as well as cross-protective antibodies. Our approach was to first define a “minimal” region within the binding domain of CIDR that determines CD36 specificity and explore its usefulness as a potential vaccine candidate. Murine polyclonal antibodies raised against the C-terminal 60 amino acids presented as a GST fusion protein are able to recognize PfEMP-1 expressed in three different unrelated parasite strains, indicating that cross-reactive antibodies were elicited. In addition, these antibodies specifically inhibited the binding of the recombinant CIDR-f to CD36. This is intriguing, as the His-tagged fragment (CIDR-Δ106) itself appears unstructured (Fig. 2D) and is unable to bind CD36 on its own (Fig. 4A). Antibodies directed against the anti-GST-CIDR-Δ106 protein are able to specifically inhibit CD36 binding of PEs while at the same time having no effect on CSA binding erythrocytes (Fig. 6B).
Further studies are now needed to characterize in structural terms the CIDRα domain and what determines its specificity for the CD36 receptor.
Acknowledgments
We are grateful to Alan Cowman for the 3D7, HB3, and FCR3 parasite clones and to Michael Lanzer for the HLEC and CHO-K1 cell lines. We also thank Kang Congbao for helping us with 1D NMR spectroscopy.
Financial support via grants from NTU (SUG 14/02), the Singapore Biomedical Research Council (03/1/21/20/291 and 02/1/22/17/043), and the Academic Research Fund (NMRC/SRG/001/2003) to the J.L. and P.P. laboratories are acknowledged.
Editor: J. F. Urban, Jr.
Footnotes
Published ahead of print on 25 February 2008.
REFERENCES
- 1.Aley, S. B., J. A. Sherwood, and R. J. Howard. 1984. Knob-positive and knob-negative Plasmodium falciparum differ in expression of a strain-specific malarial antigen on the surface of infected erythrocytes. J. Exp. Med. 1601585-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnwell, J. W., A. S. Asch, R. L. Nachman, M. Yamaya, M. Aikawa, and P. Ingravallo. 1989. A human 88-kD membrane glycoprotein (CD36) functions in vitro as a receptor for a cytoadherence ligand on Plasmodium falciparum-infected erythrocytes. J. Clin. Investig. 84765-772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baruch, D. I., X. C. Ma, B. Pasloske, R. J. Howard, and L. H. Miller. 1999. CD36 peptides that block cytoadherence define the CD36 binding region for Plasmodium falciparum-infected erythrocytes. Blood 942121-2127. [PubMed] [Google Scholar]
- 4.Baruch, D. I., X. C. Ma, H. B. Singh, X. Bi, B. L. Pasloske, and R. J. Howard. 1997. Identification of a region of PfEMP1 that mediates adherence of Plasmodium falciparum infected erythrocytes to CD36: conserved function with variant sequence. Blood 903766-3775. [PubMed] [Google Scholar]
- 5.Berendt, A. R., D. L. Simmons, J. Tansey, C. I. Newbold, and K. Marsh. 1989. Intercellular adhesion molecule-1 is an endothelial cell adhesion receptor for Plasmodium falciparum. Nature 34157-59. [DOI] [PubMed] [Google Scholar]
- 6.Buffet, P. A., B. Gamain, C. Scheidig, D. Baruch, J. D. Smith, R. Hernandez-Rivas, B. Pouvelle, S. Oishi, N. Fujii, T. Fusai, D. Parzy, L. H. Miller, J. Gysin, and A. Scherf. 1999. Plasmodium falciparum domain mediating adhesion to chondroitin sulfate A: a receptor for human placental infection. Proc. Natl. Acad. Sci. USA 9612743-12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bull, P. C., B. S. Lowe, M. Kortok, and K. Marsh. 1999. Antibody recognition of Plasmodium falciparum erythrocyte surface antigens in Kenya: evidence for rare and prevalent variants. Infect. Immun. 67733-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bull, P. C., B. S. Lowe, M. Kortok, C. S. Molyneux, C. I. Newbold, and K. Marsh. 1998. Parasite antigens on the infected red cell surface are targets for naturally acquired immunity to malaria. Nat. Med. 4358-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, Q., A. Heddini, A. Barragan, V. Fernandez, S. F. Pearce, and M. Wahlgren. 2000. The semiconserved head structure of Plasmodium falciparum erythrocyte membrane protein 1 mediates binding to multiple independent host receptors. J. Exp. Med. 1921-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cooke, B. M., C. L. Nicoll, D. I. Baruch, and R. L. Coppel. 1998. A recombinant peptide based on PfEMP-1 blocks and reverses adhesion of malaria-infected red blood cells to CD36 under flow. Mol. Microbiol. 3083-90. [DOI] [PubMed] [Google Scholar]
- 11.Florens, L., X. Liu, Y. Wang, S. Yang, O. Schwartz, M. Peglar, D. J. Carucci, J. R. Yates III, and Y. Wub. 2004. Proteomics approach reveals novel proteins on the surface of malaria-infected erythrocytes. Mol. Biochem. Parasitol. 1351-11. [DOI] [PubMed] [Google Scholar]
- 12.Gamain, B., S. Gratepanche, L. H. Miller, and D. I. Baruch. 2002. Molecular basis for the dichotomy in Plasmodium falciparum adhesion to CD36 and chondroitin sulfate A. Proc. Natl. Acad. Sci. USA 9910020-10024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gamain, B., L. H. Miller, and D. I. Baruch. 2001b. The surface variant antigens of Plasmodium falciparum contain cross-reactive epitopes. Proc. Natl. Acad. Sci. USA 982664-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gamain, B., J. D. Smith, M. Avril, D. I. Baruch, A. Scherf, J. Gysin, and L. H. Miller. 2004. Identification of a 67-amino-acid region of the Plasmodium falciparum variant surface antigen that binds chondroitin sulphate A and elicits antibodies reactive with the surface of placental isolates. Mol. Microbiol. 53445-455. [DOI] [PubMed] [Google Scholar]
- 15.Gamain, B., J. D. Smith, L. H. Miller, and D. I. Baruch. 2001a. Modifications in the CD36 binding domain of the Plasmodium falciparum variant antigen are responsible for the inability of chondroitin sulfate A adherent parasites to bind CD36. Blood 973268-3274. [DOI] [PubMed] [Google Scholar]
- 16.Gardner, M. J., N. Hall, E. Fung, O. White, M. Berriman, R. W. Hyman, J. M. Carlton, A. Pain, K. E. Nelson, S. Bowman, I. T. Paulsen, K. James, J. A. Eisen, K. Rutherford, S. L. Salzberg, A. Craig, S. Kyes, M. S. Chan, V. Nene, S. J. Shallom, B. Suh, J. Peterson, S. Angiuoli, M. Pertea, J. Allen, J. Selengut, D. Haft, M. W. Mather, A. B. Vaidya, D. M. Martin, A. H. Fairlamb, M. J. Fraunholz, D. S. Roos, S. A. Ralph, G. I. McFadden, L. M. Cummings, G. M. Subramanian, C. Mungall, J. C. Venter, D. J. Carucci, S. L. Hoffman, C. Newbold, R. W. Davis, C. M. Fraser, and B. Barrell. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419498-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giha, H. A., T. Staalsoe, D. Dodoo, I. M. Elhassan, C. Roper, G. M. Satti, D. E. Arnot, T. G. Theander, and L. Hviid. 1999. Nine-year longitudinal study of antibodies to variant antigens on the surface of Plasmodium falciparum-infected erythrocytes. Infect. Immun. 674092-4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hans, D., P. Pattnaik, A. Bhattacharyya, A. R. Shakri, S. S. Yazdani, M. Sharma, H. Choe, M. Farzan, and C. E. Chitnis. 2005. Mapping binding residues in the Plasmodium vivax domain that binds Duffy antigen during red cell invasion. Mol. Microbiol. 551423-1434. [DOI] [PubMed] [Google Scholar]
- 19.Leech, J. H., J. W. Barnwell, M. Aikawa, L. H. Miller, and R. J. Howard. 1984. Plasmodium falciparum malaria: association of knobs on the surface of infected erythrocytes with a histidine-rich protein and the erythrocyte skeleton. J. Cell Biol. 981256-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lekana Douki, J. B., B. Traore, F. T. Costa, T. Fusai, B. Pouvelle, Y. Sterkers, A. Scherf, and J. Gysin. 2002. Sequestration of Plasmodium falciparum-infected erythrocytes to chondroitin sulfate A, a receptor for maternal malaria: monoclonal antibodies against the native parasite ligand reveal pan-reactive epitopes in placental isolates. Blood 1001478-1483. [DOI] [PubMed] [Google Scholar]
- 21.Lescar, J., M. Pellegrini, H. Souchon, D. Tello, R. J. Poljak, N. Peterson, M. Greene, and P. M. Alzari. 1995. Crystal structure of a cross-reaction complex between Fab F9.13.7 and guinea fowl lysozyme. J. Biol. Chem. 27018067-18076. [DOI] [PubMed] [Google Scholar]
- 22.MacPherson, G. G., M. J. Warrell, N. J. White, S. Looareesuwan, and D. A. Warrell. 1985. Human cerebral malaria. A quantitative ultrastructural analysis of parasitized erythrocyte sequestration. Am. J. Pathol. 119385-401. [PMC free article] [PubMed] [Google Scholar]
- 23.Makobongo, M. O., B. Keegan, C. A. Long, and L. H. Miller. 2006. Immunization of Aotus monkeys with recombinant cysteine-rich interdomain region 1 alpha protects against severe disease during Plasmodium falciparum reinfection. J. Infect. Dis. 193731-740. [DOI] [PubMed] [Google Scholar]
- 24.Miller, L. H. 1969. Distribution of mature trophozoites and schizonts of Plasmodium falciparum in the organs of Aotus trivirgatus, the night monkey. Am. J. Trop. Med. Hyg. 18860-865. [DOI] [PubMed] [Google Scholar]
- 25.Moll, K., F. Pettersson, A. M. Vogt, C. Jonsson, N. Rasti, S. Ahuja, M. Spangberg, O. Mercereau-Puijalon, D. E. Arnot, M. Wahlgren, and Q. Chen. 2007. Generation of cross-protective antibodies against Plasmodium falciparum sequestration by immunization with an erythrocyte membrane protein 1-Duffy binding-like 1 alpha domain. Infect. Immun. 75211-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novotny, J., R. Bruccoleri, J. Newell, D. Murphy, E. Haber, and M. Karplus. 1983. Molecular anatomy of the antibody binding site. J. Biol. Chem. 25814433-14437. [PubMed] [Google Scholar]
- 27.Page, R., W. Peti, I. A. Wilson, R. C. Stevens, and K. Wuthrich. 2005. NMR screening and crystal quality of bacterially expressed prokaryotic and eukaryotic proteins in a structural genomics pipeline. Proc. Natl. Acad. Sci. USA 1021901-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pouvelle, B., P. Meyer, C. Robert, L. Bardel, and J. Gysin. 1997. Chondroitin-4-sulfate impairs in vitro and in vivo cytoadherence of Plasmodium falciparum infected erythrocytes. Mol. Med. 3508-518. [PMC free article] [PubMed] [Google Scholar]
- 29.Reeder, J. C., and G. V. Brown. 1996. Antigenic variation and immune evasion in Plasmodium falciparum malaria. Immunol. Cell Biol. 74546-554. [DOI] [PubMed] [Google Scholar]
- 30.Roberts, D. D., J. A. Sherwood, S. L. Spitalnik, L. J. Panton, R. J. Howard, V. M. Dixit, W. A. Frazier, L. H. Miller, and V. Ginsburg. 1985. Thrombospondin binds falciparum malaria parasitized erythrocytes and may mediate cytoadherence. Nature 31864-66. [DOI] [PubMed] [Google Scholar]
- 31.Robinson, B. A., T. L. Welch, and J. D. Smith. 2003. Widespread functional specialization of Plasmodium falciparum erythrocyte membrane protein 1 family members to bind CD36 analysed across a parasite genome. Mol. Microbiol. 471265-1278. [DOI] [PubMed] [Google Scholar]
- 32.Rogerson, S. J., S. C. Chaiyaroj, K. Ng, J. C. Reeder, and G. V. Brown. 1995. Chondroitin sulfate A is a cell surface receptor for Plasmodium falciparum-infected erythrocytes. J. Exp. Med. 18215-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh, S. K., R. Hora, H. Belrhali, C. E. Chitnis, and A. Sharma. 2006. Structural basis for Duffy recognition by the malaria parasite Duffy-binding-like domain. Nature 439741-744. [DOI] [PubMed] [Google Scholar]
- 34.Smith, J. D., C. E. Chitnis, A. G. Craig, D. J. Roberts, D. E. Hudson-Taylor, D. S. Peterson, R. Pinches, C. I. Newbold, and L. H. Miller. 1995. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell 82101-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith, J. D., A. G. Craig, N. Kriek, D. Hudson-Taylor, S. Kyes, T. Fagen, R. Pinches, D. I. Baruch, C. I. Newbold, and L. H. Miller. 2000b. Identification of a Plasmodium falciparum intercellular adhesion molecule-1 binding domain: a parasite adhesion trait implicated in cerebral malaria. Proc. Natl. Acad. Sci. USA 971766-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith, J. D., G. Subramanian, B. Gamain, D. I. Baruch, and L. H. Miller. 2000a. Classification of adhesive domains in the Plasmodium falciparum erythrocyte membrane protein 1 family. Mol. Biochem. Parasitol. 110293-310. [DOI] [PubMed] [Google Scholar]
- 37.Su, X. Z., V. M. Heatwole, S. P. Wertheimer, F. Guinet, J. A. Herrfeldt, D. S. Peterson, J. A. Ravetch, and T. E. Wellems. 1995. The large diverse gene family var encodes proteins involved in cytoadherence and antigenic variation of Plasmodium falciparum-infected erythrocytes. Cell 8289-100. [DOI] [PubMed] [Google Scholar]
- 38.Tolia, N. H., E. J. Enemark, B. K. Sim, and L. Joshua-Tor. 2005. Structural basis for the EBA-175 erythrocyte invasion pathway of the malaria parasite Plasmodium falciparum. Cell 122183-193. [DOI] [PubMed] [Google Scholar]
- 39.Trossaert, M., A. Dieye, Y. Dieye, and J. L. Sarthou. 1991. Cytoadherence of Plasmodium falciparum and complications of malaria. Dakar Med. 36192-197. [PubMed] [Google Scholar]
- 40.Yipp, B. G., D. I. Baruch, C. Brady, A. G. Murray, S. Looareesuwan, P. Kubes, and M. Ho. 2003. Recombinant PfEMP1 peptide inhibits and reverses cytoadherence of clinical Plasmodium falciparum isolates in vivo. Blood 101331-337. [DOI] [PubMed] [Google Scholar]