Abstract
The narKGHJI operon that comprises putative nitrate/nitrite transporter (narK) and nitrate reductase (narGHJI) genes is required for the anaerobic growth of Corynebacterium glutamicum with nitrate as a terminal electron acceptor. In this study, we identified a gene, arnR, which encodes a transcriptional regulator that represses the expression of the narKGHJI operon in C. glutamicum cells under aerobic conditions. Disruption of arnR induced nitrate reductase activities of C. glutamicum cells and increased narKGHJI mRNA levels under aerobic growth conditions. DNA microarray analyses revealed that besides the narKGHJI operon, the hmp gene, which encodes flavohemoglobin, is negatively regulated by ArnR under aerobic conditions. Promoter-reporter assays indicated that arnR gene expression was positively autoregulated by its gene product, ArnR, under both aerobic and anaerobic conditions. Electrophoretic mobility shift assay experiments showed that purified hexahistidyl-tagged ArnR protein specifically binds to promoter regions of the narKGHJI operon and the hmp and arnR genes. A consensus sequence, TA(A/T)TTAA(A/T)TA, found in the promoter regions of these genes was demonstrated to be involved in the binding of ArnR. Effects on LacZ activity by deletion of the ArnR binding sites within the promoter regions fused to the reporter gene were consistent with the view that the expression of the narKGHJI operon is repressed by the ArnR protein under aerobic conditions, whereas the expression of the arnR gene is autoinduced by ArnR.
Nitrate respiration is an important physiological process that allows bacteria to generate sufficient energy to permit anaerobic growth (21, 49). The genetics and biochemistry of nitrate respiration have been intensively studied in Escherichia coli (4, 21). During nitrate respiration, nitrate is reduced to nitrite in the cytoplasm by a membrane-bound nitrate reductase. The three subunits of the nitrate reductase complex (NarGHI) and a fourth polypeptide required for the assembly of the complex (NarJ) are encoded by the narGHJI operon. In addition, in the E. coli genome, a gene, narK, that encodes a nitrate/nitrite transporter is located upstream of the operon and in the same orientation but constitutes a distinct transcription unit (4, 7).
The expression of E. coli narGHJI and narK is activated by the Fnr protein in response to anaerobiosis (4). Fnr is a global transcription regulator that activates the expression of genes encoding many of the enzymes required for the anaerobic metabolism of E. coli (10, 37). The activation of Fnr requires the formation of an oxygen-sensitive Fe-S cluster (9, 13), and disassembly of the labile Fe-S cluster in the presence of oxygen prevents the transcriptional activation of the target genes (44). E. coli Fnr constitutes the paradigm of oxygen-sensing regulators, which have been found in a variety of gram-negative and a few gram-positive bacteria (17, 35, 36, 49). Notably, in Bacillus subtilis, expression of the narGHJI operon and expression of the narK-fnr operon are activated by Fnr in response to anaerobiosis as in E. coli (8, 25, 32). However, unlike in E. coli, where fnr expression is almost constitutive (38), the transcription of the B. subtilis fnr gene is highly induced by anaerobiosis via the ResD-ResE two-component system (24, 26). The latter system constitutes a major regulatory switch for B. subtilis to adapt to anaerobiosis (41, 47). Furthermore, it is also worth noting that E. coli nar gene expression is up-regulated in response to nitrate via a NarX-NarL two-component system (39, 40). Nitrate induction of narK and narGHJI similarly occurs under anaerobic conditions in B. subtilis (8, 32); however, the sensing mechanism remains to be elucidated.
Corynebacterium glutamicum is a nonpathogenic gram-positive soil bacterium. This high-GC-content microorganism has been widely used for the industrial production of various amino acids, nucleic acids, and commodity chemicals (11, 16, 43). Recent studies showed that C. glutamicum grows anaerobically using nitrate as a terminal electron acceptor (28, 42). This bacterium consumes nitrate and excretes nitrite as the main end product of nitrate reduction during anaerobic growth. This property is attributed to the presence of a narKGHJI operon with high similarity to the E. coli narK gene and narGHJI operon. Moreover, we observed that the expression of the narKGHJI operon is induced by anaerobiosis and additionally by the presence of nitrate (28). This expression pattern is similar to that of the narK gene and narGHJI operon in E. coli and B. subtilis (4, 8, 32). Notably, the expression of C. glutamicum narKGHJI is negatively regulated by the AraC-type regulator RipA under conditions of iron limitation (46), but the relationship between RipA and anaerobiosis- or nitrate-mediated induction of the operon remains unclear.
In the present study, we identified a gene, cgR1264, whose product (CgR1264) is a novel transcriptional regulator of the narKGHJI operon in C. glutamicum. We showed that CgR1264 represses the expression of the narKGHJI operon under aerobic conditions. This activity contrasts with that of Fnr, which anaerobically activates the transcription of nitrate reductase genes in E. coli and B. subtilis (4, 8, 32). Therefore, we designated this protein ArnR (aerobic repressor of nitrate reductase R). ArnR was also shown to directly repress the expression of the hmp gene, encoding flavohemoglobin, and to positively regulate its own gene expression. Furthermore, a consensus sequence found within the promoter regions of the genes regulated by ArnR was shown to be involved in the aerobic repression of expression of the operon and in the constitutive activation of the expression of the arnR gene.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Strains used in this study are listed in Table 1. C. glutamicum R (48) and its derivative strains were precultured at 33°C overnight in nutrient-rich medium (A medium) containing (per liter) 2 g urea, 2 g yeast extract, 7 g Casamino Acids, 7 g (NH4)2SO4, 0.5 g KH2PO4, 0.5 g K2HPO4, 0.5 g MgSO4·7H2O, 6 mg FeSO4·7H2O, 4.2 mg MnSO4·H2O, 0.2 mg biotin, and 0.2 mg thiamine with 1% (wt/vol) glucose. Precultures were harvested by centrifugation; the resulting cell precipitates were subsequently washed once with BT minimal medium (A medium without yeast extract and Casamino Acids). For aerobic cultivation, washed cells were grown at 33°C in 100 ml BT medium containing 0.5% (wt/vol) glucose in a 500-ml flask with vigorous shaking (200 rpm). Potassium nitrate was added to a concentration of 30 mM where necessary. For anaerobic cultivation, anaerobic BT medium was prepared in advance by bubbling argon for 5 min; cells from the same batch as those used for aerobic cultivation were subsequently transferred into anaerobic BT medium in an anaerobic chamber (Coy Laboratory Products, Grass Lake, MI), where the gas composition was kept constant at 95% nitrogen and 5% hydrogen. Cells were grown at 33°C in 50 ml BT medium containing 0.5% (wt/vol) glucose in a 50-ml medium bottle with gentle stirring. Potassium nitrate was added to a concentration of 30 mM when necessary. Kanamycin was used at 50 μg/ml for the cultivation of the ΔarnR strain. Chloramphenicol was used at 5 μg/ml for the cultivation of R orΔarnR strains carrying various promoter-lacZ plasmids.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| C. glutamicum R | Wild type | 48 |
| ΔarnR | R arnR::Kmr | This work |
| E. coli | ||
| JM109 | endA1 gyrA96 thi hsdR17 supE44 relA1 Δ(lac-proAB) recA1 F′[traD36 proAB+lacIqlacZΔM15] | TaKaRa |
| JM110 | dam dcm supE44 hsdR17 thi leu rpsL acy galK galT ara tonA thr tsx Δ(lac-proAB)/F′ [traD36 proAB+lacIqlacZΔM15] | 34 |
| BL21(DE3) | ompT hsdSB(rB− mB−) gal dcm (DE3) | Novagen |
| Plasmids | ||
| pHSG398 | Cmr; cloning vector | TaKaRa |
| pUC4K | Apr Kmr; source of Kmr cartridge | Pharmacia |
| pMC1871 | Tcr; lacZ fusion vector | Pharmacia |
| pCRB1 | Cmr; α-lac multicloning site; E. coli-Corynebacterium sp. shuttle vector | 27 |
| pET-21a(+) | Apr; vector for overexpression of genes in E. coli, adding a C-terminal hexahistidine affinity tag to the synthesized protein | Novagen |
| pCRB200 | Cmr; lacZ reporter assay vector | This work |
| pCRB601 | Cmr; pHSG398 with a 4.9-kb BamHI-XbaI PCR fragment containing the C. glutamicum R arnR gene | This work |
| pCRB602 | Cmr; a 1.0-kb Kmr cartridge inserted into the pCRB601 BglII-SpeI site | This work |
| pCRB603 | Cmr; a 281-bp narKGHJI promoter fragment inserted into the pCRB200 SmaI site in the forward direction | This work |
| pCRB604 | Cmr; a 254-bp arnR promoter fragment inserted into the pCRB200 SmaI site in the forward direction | This work |
| pCRB605 | Cmr; pCRB603 lacking the ArnR binding site | This work |
| pCRB606 | Cmr; pCRB604 lacking the ArnR binding site | This work |
| pCRB607 | Apr; the C. glutamicum R arnR coding region inserted into the pET-21a(+) NdeI-XhoI site | This work |
E. coli JM109 was used as a host for all genetic manipulations. E. coli BL21(DE3) (Novagen, Madison, WI) was used for protein expression. E. coli strains were cultivated at 37°C in LB medium supplemented, when necessary, with 50 μg/ml ampicillin, kanamycin, or chloramphenicol.
Construction of an arnR-deficient strain.
Plasmids used in this study are listed in Table 1. Primers used in this study are listed in Table S1 in the supplemental material. Plasmid pCRB601 was obtained by PCR by amplifying the arnR gene of C. glutamicum R using primers arnR-f and arnR-r, digesting the product with BamHI and XbaI, and inserting the resulting digest into the BamHI and XbaI sites of pHSG398. Plasmid pCRB602 was constructed from pCRB601 by inverse PCR amplification using primers arnRinv-f and arnRinv-r and digesting the product with BglII and SpeI, followed by religating with a BglII- and SpeI-digested kanamycin cartridge. The latter cartridge was obtained by PCR amplifying the Kmr region of pUC4K with primers kan-f and kan-r. The resulting plasmid, pCRB602, was used to transform C. glutamicum R by electroporation to generate an arnR-deficient strain according to a method using nonmethylated DNA isolated from E. coli JM110 as reported previously (45).
Construction of promoter-lacZ fusions.
Plasmids pCRB603 to pCRB606 (Table 1) for β-galactosidase assays were constructed as follows. LacZ reporter assay vector pCRB200 was constructed from pCRB1 by inverse PCR amplification using primers PLSV-f and PLSV-r and digesting the product with PstI, followed by religation with a PstI-digested lacZ cartridge lacking a translation start codon. The latter cartridge was obtained by cutting out the lacZ region of pMC1871 with PstI digestion. A 281-bp DNA fragment covering the intergenic region between the mog and narK genes and a 253-bp DNA fragment covering the intergenic region between the narI and arnR genes were amplified by PCR using C. glutamicum R genomic DNA as a template and the following primer pairs: narKlacZ-f and narKlacZ-r and arnRlacZ-f and arnRlacZ-r, respectively. The two PCR products were digested with SmaI and inserted into the SmaI site of pCRB200, resulting in plasmids pCRB603 and pCRB604, respectively. The direction of the inserts was confirmed by DNA sequencing. Transformants harboring the recombinant plasmid with the insert in the forward or reverse direction appeared as pale blue or white colonies on A medium agar plates containing 5 μg/ml of chloramphenicol and 100 μg/ml of 5-bromo-4-chloro-β-d-galactopyranoside (X-Gal). Deletion of the putative ArnR binding site (ARS) within the narK promoter region was performed by inverse PCR using pCRB603 as a template and primers narKlacZinv-f and narKlacZinv-r. Similarly, the putative ARS within the arnR promoter region was deleted by inverse PCR using the primer pair arnRlacZinv-f and arnRlacZinv-r and pCRB604 as template. The resulting PCR products were phosphorylated with T4 polynucleotide kinase (TaKaRa Bio, Otsu, Japan) and self-ligated to generate pCRB605 and pCRB606, respectively. The resulting plasmids, pCRB603 to pCRB606, were introduced into C. glutamicum R or its recombinant derivative ΔarnR strain according to a method using nonmethylated DNA isolated from E. coli JM110 (45).
QRT-PCR analysis.
C. glutamicum R wild-type and ΔarnR cells were grown to mid-log phase aerobically without nitrate or anaerobically with nitrate. Cells were harvested from aerobic and anaerobic cultures. Total RNAs were subsequently extracted from these samples with an RNeasy Mini kit (Qiagen, MD) according to the manufacturer's instructions. Residual DNA was removed by treatment with RQ1 RNase-free DNase (Promega, Madison, WI). Quantitative reverse transcription-PCR (QRT-PCR) analyses were performed using a Power Sybr green PCR master mix protocol (Applied Biosystems, Warrington, United Kingdom), according to the manufacturer's instructions, with an ABI Prism 7000 sequence detection system (Applied Biosystems). Individual target genes were amplified with the primers listed in Table S1 in the supplemental material. Fifty nanograms of total RNA per well was incubated for 30 min at 50°C for reverse transcription, heated for 10 min at 95°C for initial PCR activation, and thereafter amplified for 40 cycles, with each cycle consisting of denaturation at 95°C for 15 s and annealing and extension at 60°C for 1 min. To account for any nucleic acid contamination, negative controls without reverse transcriptase or template RNA were run. Reverse transcription-PCR for each gene was performed with three independent RNA samples.
DNA microarray analysis.
C. glutamicum R wild-type and ΔarnR strains were cultivated in BT medium under aerobic conditions without nitrate, and their total RNAs were extracted with an RNeasy Mini kit (Qiagen, MD) according to the manufacturer's instructions. Residual DNA was removed by treating the preparation with RQ1 RNase-free DNase (Promega, Madison, WI). Preparation of the C. glutamicum R whole-genome DNA microarray and fluorescently labeled cDNA from the total RNA and microarray hybridization, washing, and data analysis were performed as described previously (12). The resultant raw data were shown as average mRNA ratios of a total of six signals for each gene: duplicate spots on three different slides of three replicate RNA samples from three independent experiments. Genes that showed significantly altered mRNA levels (P value of <0.01 by a Student's t test) by a factor of 2 or more were determined.
Overproduction and purification of the hexahistidyl-tagged ArnR protein.
In order to overexpress and purify the hexahistidyl-tagged ArnR (ArnR-His6) protein, the arnR open reading frame was amplified by PCR using the primer pair arnRorf-f and arnRorf-r. The PCR product was digested with NdeI and XhoI, and the digest was inserted into the NdeI and XhoI sites of pET-21a(+) (Novagen, Madison, WI). The resulting plasmid, pCRB607, was introduced into E. coli BL21(DE3) by electroporation.
E. coli BL21(DE3) carrying pCRB607 was grown in 100 ml of LB medium containing ampicillin at 37°C to an optical density at 610 nm of approximately 0.5. Expression of ArnR-His6 protein was induced by the addition of isopropyl-β-d-thiogalactopyranoside (TaKaRa Bio, Otsu, Japan). After 4 h of incubation at 37°C, cells were harvested by centrifugation. The ArnR-His6 protein was purified under native conditions with a QIAexpress Ni-NTA Fast Start kit (Qiagen) according to the manufacturer's protocol. The protein concentration was determined with a protein assay (Japan Bio-Rad Laboratories, Tokyo, Japan).
EMSA.
A series of the upstream fragments of narKGHJI and arnR (Pnar1 to Pnar5 and ParnR1 to ParnR4) and the hmp promoter fragment (Phmp) were amplified by PCR using the following primer pairs: Pnar1-f and Pnar-r for Pnar1, Pnar2-f and Pnar-r for Pnar2, Pnar3-f and Pnar-r for Pnar3, Pnar4-f and Pnar-r for Pnar4, Pnar5-f and Pnar-r for Pnar5, ParnR1-f and ParnR-r for ParnR1, ParnR2-f and ParnR-r for ParnR2, ParnR3-f and ParnR-r for ParnR3, ParnR4-f and ParnR-r for ParnR4, and Phmp-f and Phmp-r for Phmp. Fragments (30 bp) containing wild-type or mutated ARS were prepared by annealing primer pairs wt-f and wt-r for the wild-type fragment, m1-f and m1-r for the M1 fragment, m2-f and m2-r for the M2 fragment, and m3-f and m3-r for the M3 fragment. The resulting double-stranded fragments were used in electrophoretic mobility shift assays (EMSAs).
Binding reactions were performed using a solution containing 10 mM Tris-HCl (pH 7.4), 50 mM KCl, 0.1 mM dithiothreitol, and 0.1 mM EDTA. A total of 12.5 nM specific or nonspecific DNA fragment and 0 to 800 nM (0 to 64 molar excess) of purified ArnR-His6 protein were added to a final volume of 10 μl and incubated at room temperature for 15 min. The samples were immediately loaded onto a 5 or 10% nondenaturing polyacrylamide gel and electrophoresed at 200 V in 0.5× Tris-borate-EDTA buffer (44.5 mM Tris base, 44.5 mM boric acid, and 1 mM EDTA). The gel was stained using an EMSA kit (Invitrogen).
Primer extension analysis.
IRD700-labeled primer IRD-hmp1 was designed as shown in Table S1 in the supplemental material. Ten micrograms of total RNA and 1.5 pmol of primer were mixed and annealed at 60°C for 20 min and at room temperature for 5 min. cDNA was synthesized at 42°C for 30 min using avian myeloblastosis virus reverse transcriptase (Promega, Madison, WI). The reaction was terminated by adding EDTA (pH 8.0) at a final concentration of 250 mM and DNase-free RNase A at a final concentration of 3 ng/μl to the mixture to trigger the degradation of the RNA templates. The resulting cDNA was treated by phenol-chloroform extraction and ethanol precipitation. Upon centrifugation, the precipitated DNA pellet was resuspended in IR2 stop solution (Li-Cor, NE). The primer extension products were treated at 95°C for 5 min, placed on ice for 5 min, and separated on a 5.5% KB Plus gel matrix (Li-Cor, NE) using a Li-Cor 4300 DNA analyzer. The migration position of the primer extension product was determined by comparing a sequencing ladder generated from a DNA fragment corresponding to the same chromosomal region using the same primers and a DYEnamic direct cycle sequencing kit with 7-deaza-dGTP (Amersham Biosciences, NJ).
DNase I footprinting.
The narK promoter DNA fragment was amplified by using primer pair PnarK1-f and IRD-narK1 (see Table S1 in the supplemental material). An IRD700-labeled DNA fragment (2 nM) was mixed with purified ArnR in a total volume of 100 μl of binding buffer (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 3 mM MgCl2, 5 mM CaCl2, 0.1 mM EDTA, 0.1 mM dithiothreitol, 5% [wt/vol] glycerol, and 50 μM bovine serum albumin). The mixture was incubated for 10 min at 33°C and then placed at 25°C for 5 min. DNase I (TaKaRa Bio, Otsu, Japan) was added at 0.1 U, and incubation was continued for 1 min at 25°C. The digestion was stopped with 350 μl ice-cold stop solution (324 μl of 99% [vol/vol] ethanol, 1 μl of salmon sperm DNA [10 μg/μl], 25 μl of saturated ammonium acetate solution), and DNA was precipitated overnight at −20°C. The precipitated DNA pellet was resuspended in IR2 stop solution (Li-Cor, NE). The dissolved sample was separated as described above for primer extension analysis.
β-Galactosidase assay.
Toluene treatment and β-galactosidase assays using o-nitrophenyl-β-d-galactopyranoside were performed as previously described (20). The values presented are the means of at least three independent experiments.
Analytical methods.
Cell growth was monitored by measuring the absorbance at 610 nm using a UV-visible DU 730 spectrophotometer (Beckman Coulter, CA). Nitrite concentration was determined by using an NO2/NO3 Assay Kit-C II (Colorimetric) and a Griess reagent kit (Dojindo, Kumamoto, Japan). Protein concentrations were measured using a Bio-Rad protein assay kit (Japan Bio-Rad Laboratories, Tokyo, Japan) with bovine serum albumin as the standard (5). The UV-visible light spectrum of the purified ArnR was recorded using a UV-visible DU730 spectrophotometer (Beckman Coulter, CA).
RESULTS
Identification of a transcriptional repressor of expression of the narKGHJI operon.
Previous studies showed that the expression of the narKGHJI operon is induced by anaerobiosis and nitrate in C. glutamicum (28). In order to unravel the genetic regulation mechanisms of anaerobic growth in C. glutamicum, we focused our initial efforts on arnR, a gene that is located immediately downstream of the narKGHJI operon (48). The product of this gene (ArnR) consists of 230 amino acids. Although the deduced ArnR protein does not exhibit any significant similarity to any other protein of known function as determined by BLAST searches, the protein was previously reported to be classified as a putative transcriptional regulator that possesses a winged-helix-type DNA-binding domain in the amino-terminal region (6). The amino-terminal DNA-binding domain of ArnR showed no significant similarity to the carboxyl-terminally-located DNA-binding domains of Fnr-type regulators involved in nar gene expression in E. coli and B. subtilis (4, 8, 32). A homology search revealed that ArnR-homologous proteins are present in a discrete number of Actinobacteria, such as Corynebacterium efficiens (CE1292 [62% amino acid sequence identity]), Corynebacterium diphtheriae (DIP0494 [37% identity]), Mycobacterium tuberculosis (Rv2621c [28% identity]), and Nocardioides sp. (Noca_1344 [28% identity]). As in the C. glutamicum chromosome, in the C. efficiens CE1292 chromosome, this gene is located downstream of and adjacent to the narKGHJI gene cluster and in the same orientation, whereas in the Noca_1344 genome of Nocardioides sp., it is adjacent to, but upstream of, the nitrate reductase gene cluster and in the reverse orientation.
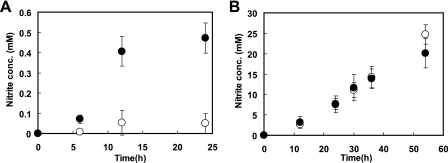
To examine the role of ArnR in the regulation of the expression of the narKGHJI operon, we constructed an arnR-deficient strain (ΔarnR). Both ΔarnR and wild-type R strains were grown in BT minimal medium containing 0.5% (wt/vol) glucose as a sole carbon source under aerobic or anaerobic growth conditions. Under anaerobic conditions, 30 mM potassium nitrate was added to serve as a terminal electron acceptor. To assess the capability of nitrate reduction in C. glutamicum strains, nitrite production was monitored in the aerobically and anaerobically growing cultures containing nitrate. ΔarnR did not show a significant growth defect under aerobic and anaerobic conditions (data not shown). Under conditions of aerobic growth in the presence of nitrate, whereas nitrite production was hardly observed in the wild-type culture, a much higher nitrite production rate was detected in the ΔarnR culture (Fig. 1A). On the other hand, there was no significant difference in the production rates of nitrite between the two strains during anaerobic growth with nitrate (Fig. 1B).
FIG. 1.
Changing concentrations of nitrite in C. glutamicum R wild-type (open circles) and ΔarnR (closed circles) cultures during aerobic (A) or anaerobic (B) growth in the presence of 30 mM of nitrate. The means ± standard deviations (error bars) of three independent experiments are shown.
To further understand this phenotypical change exhibited by ΔarnR cells incubated under aerobic conditions, we examined whether a disruption of arnR affects the transcriptional levels of the narK and narG genes used as representative expression level markers of the narKGHJI operon. QRT-PCR analyses revealed that mRNA levels of both narK and narG in ΔarnR cells were approximately eightfold higher than those in wild-type cells under aerobic growth conditions (Table 2). This observation, together with the enhanced capability of nitrate reduction in aerobic ΔarnR cultures, suggested that C. glutamicum ArnR is involved in repressing the expression of the narKGHJI operon under aerobic conditions. In contrast, the mRNA levels of these two genes were induced under anaerobic conditions with nitrate in ΔarnR and wild-type cells, although the levels in the ΔarnR cells were slightly lower than those in the wild type. This observation suggested the presence of additional regulators acting at the level of the control of narKGHJI expression.
TABLE 2.
Influence of the arnR deletion on transcription of the narK and narG genes
| Target gene and strain | Relative mRNA level ± SDa
|
|
|---|---|---|
| Aerobic | Anaerobic with nitrate | |
| narK | ||
| R (wild type) | 1 | 36.1 ± 4.6 |
| ΔarnR | 7.9 ± 1.5 | 26.1 ± 6.4 |
| narG | ||
| R (wild type) | 1 | 24.3 ± 1.9 |
| ΔarnR | 7.7 ± 1.4 | 17.7 ± 2.8 |
Reported values represent the means ± standard deviations of three independent experiments.
The hmp gene, encoding flavohemoglobin, is an additional target of ArnR.
We next investigated whether C. glutamicum ArnR regulates multiple genes, including genes possibly related to anaerobic metabolism, by DNA microarray analyses using C. glutamicum R wild-type and ΔarnR cells. Total RNA was extracted from the two strains aerobically grown in BT minimal medium without nitrate. Table 3 indicates genes whose average mRNA ratios (ΔarnR/wild type) were significantly (P < 0.01) altered by a factor of 2 or more, as calculated from three independent experiments. Enhanced expression levels of the nar genes in ΔarnR cells were observed in DNA microarray experiments; this is consistent with the result obtained by QRT-PCR analyses, as shown in Table 2. Additionally, a gene, hmp, encoding flavohemoglobin, showed a higher expression level in the ΔarnR strain than in the wild type, and the expression pattern was confirmed by QRT-PCR analyses. Notably, hmp was strongly induced under anaerobic conditions in the presence of nitrate (approximately 400-fold [mRNA expression levels of 395 ± 86 for the wild type and 341 ± 34 for the ΔarnR strain versus mRNA expression levels of 1 for the wild type and 79.4 ± 7.8 for the ΔarnR strain under aerobic conditions]). However, the deletion of arnR did not affect the anaerobic induction of hmp expression as in the case of the narKGHJI operon.
TABLE 3.
Genes showing altered expression in the C. glutamicum ΔarnR strain relative to the wild type under aerobic conditions
| Locus | Gene | Annotation | DNA microarray ratioa |
|---|---|---|---|
| CgR1265 | narI | Nitrate reductase, γ subunit | 3.1 |
| CgR1266 | narJ | Nitrate reductase, δ subunit | 2.7 |
| CgR1267 | narH | Nitrate reductase, β subunit | 3.0 |
| CgR1268 | narG | Nitrate reductase, α subunit | 5.6 |
| CgR1269 | narK | Putative nitrate/nitrite transporter | 6.3 |
| CgR2724 | hmp | Flavohemoglobin | 29.1 |
Genes whose average mRNA ratios (ΔarnR/wild type) were altered by >2.0-fold (P value of <0.01) in six hybridization signals for each gene (duplicate spots on three different slides of three replicate RNA samples from three independent experiments).
Autoregulation of arnR.
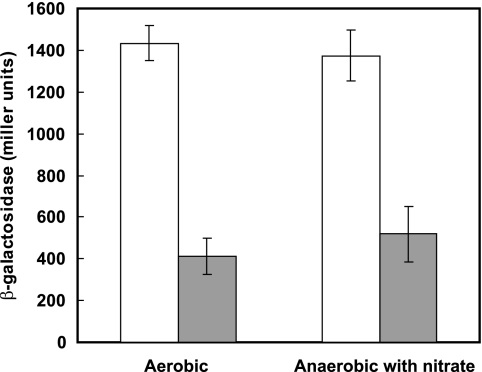
We examined whether the expression of arnR is regulated by its own gene product via an arnR promoter activity assay in C. glutamicum R wild-type or ΔarnR cells using the lacZ reporter system. The intergenic region between arnR and narI was fused to the lacZ reporter gene and borne by plasmid pCRB604. As shown in Fig. 2, the β-galactosidase activity of the wild-type strain carrying pCRB604 was shown to be almost constitutive under aerobic and anaerobic growth conditions. The ΔarnR strain carrying pCRB604 showed lower β-galactosidase activity than did wild-type cells carrying the same plasmid under both conditions, suggesting that arnR expression is positively autoregulated by its own gene product. On the other hand, reverse transcription-PCR analysis using primers corresponding to the 184-bp arnR-narI intergenic region showed that arnR is also transcribed with the narKGHJI operon from the narK promoter as well as from its own promoter under anaerobic conditions with nitrate (data not shown), although a putative stem-loop structure is found immediately downstream of the narI gene (28).
FIG. 2.
β-Galactosidase activities (Miller units) of C. glutamicum R wild-type (white bars) and ΔarnR (gray bars) strains harboring an arnR-lacZ fusion plasmid (pCRB604) under aerobic conditions and anaerobic conditions with nitrate. The means ± standard deviations (error bars) of three independent experiments are shown.
Binding of purified ArnR protein to promoter regions of the narKGHJI, hmp, and arnR genes.
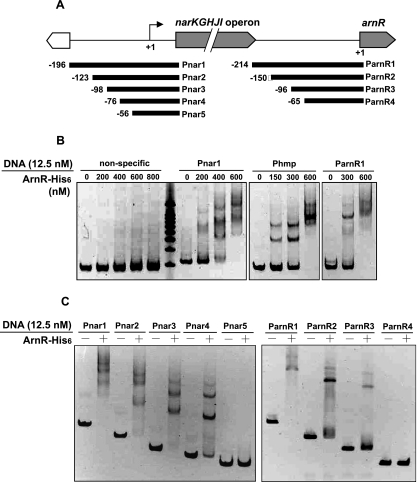
To investigate whether ArnR directly regulates the transcription of the narKGHJI operon, hmp, and arnR (its own gene), we performed EMSAs to test the specific binding of ArnR to promoter regions of these genes. ArnR was overexpressed as a hexahistidyl-tagged fusion protein (ArnR-His6) in E. coli BL21(DE3) and purified to apparent homogeneity by affinity chromatography. The ArnR-His6 protein preparation was verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis, and the presence of a protein of the expected size (25.1 kDa) was confirmed (data not shown). Subsequently, we prepared a 281-bp specific DNA probe, Pnar1, covering the intergenic region between narK and its adjacent mog genes (Fig. 3A) and, as a negative control, a nonspecific 278-bp DNA fragment corresponding to the intergenic region between the narK and narG genes. Multiple Pnar1-ArnR complexes were observed at a (protein/DNA) molar excess of 16 to 48, whereas no nonspecific DNA fragment band was shifted when ArnR was added at molar excesses of up to 64 (Fig. 3B). Likewise, we examined the binding of ArnR to the promoter regions of the hmp gene and of its own gene. A 274-bp promoter fragment of hmp, Phmp (not shown), and a 253-bp promoter fragment of arnR, ParnR1, covering the intergenic region between the narI and arnR genes (Fig. 3A), were completely shifted by ArnR at a molar excess of 48 (Fig. 3B). Multiple Phmp-ArnR and ParnR1-ArnR complexes were observed. These in vitro binding assays, together with phenotypical changes exhibited by the arnR-disrupted strain, revealed that ArnR directly regulates the expression of the narKGHJI operon, of the hmp gene, and of its own gene.
FIG. 3.
(A) Genomic locus of promoter regions of the narKGHJI operon and the arnR gene and various 5′ deletion DNA probes of their promoter regions used for EMSAs. Numbers adjacent to the fragments represent nucleotide positions relative to the narKGHJI transcriptional start site or arnR translation start codon. (B) EMSAs using 12.5 nM nonspecific probe, Pnar1, Phmp, or ParnR1 as a DNA probe and 0 to 800 nM purified ArnR-His6 protein at a molar excess (protein/DNA) of 0 to 64 for nonspecific probe and 0 to 48 for Pnar1, Phmp, and ParnR1. (C) EMSAs using a series of 5′ deletion DNA probes (12.5 nM) (Pnar1 to Pnar5 and ParnR1 to ParnR4) of the promoter regions of narKGHJI or arnR and the ArnR-His6 protein (0 or 600 nM) at a molar excess of 0 (minus sign) or 48 (plus sign).
ArnR consensus binding sequence.
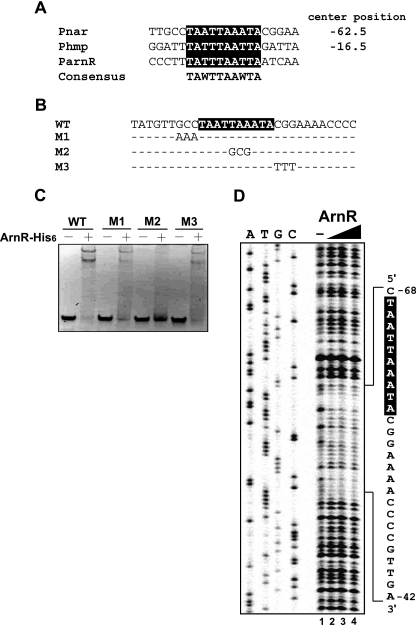
To determine the ArnR binding site present within the narKGHJI promoter region, we performed EMSAs using a set of 5′ deletion fragments (Pnar2 to Pnar5) of the full-length Pnar1 fragment (Fig. 3A). As shown in Fig. 3C, fragments Pnar1 to Pnar4 were shifted by ArnR at a molar excess of 48, whereas the Pnar5 fragment was not shifted by ArnR at the same molar excess. This indicates that the region between positions −76 and −57 upstream of the transcriptional start site is the most important one for ArnR binding. A similar experiment was performed using fragments ParnR1 to ParnR4 to examine the ArnR binding site within the arnR promoter region (Fig. 3A). Fragments ParnR1 to ParnR3 were shifted by ArnR at a molar excess of 48, whereas fragment ParnR4 was not shifted by ArnR at the same molar excess (Fig. 3C). This shows that the ArnR binding site is likely located between positions −101 and −66 upstream of the translation start codon. Based on these two narrowed promoter regions, we inferred the ARS consensus sequence. This sequence consists of a 10-bp imperfect palindrome: TA(A/T)TTAA(A/T)TA. A similar sequence is also found within the hmp promoter region (Fig. 4A). The ARS consensus sequence is centered at positions −62.5 and −16.5 relative to the previously determined transcriptional start site within the narKGHJI promoter region (28) and to the transcriptional start site of hmp newly determined by primer extension analysis, respectively. However, we detected no band for the transcriptional start site of arnR by primer extension analysis, probably due to a low expression level and/or instability of the transcript. The ARS sequence was located at positions −85 to −76 from the translation start codon within the arnR promoter region.
FIG. 4.
(A) Deduced ArnR binding sites (black box) within the promoter regions of narKGHJI, hmp, and arnR are aligned. The consensus sequence (TAWTTAAWTA) is shown below the alignment (W indicates A or T). The center positions (positions −62.5 and −16.5) of the proposed binding site relative to the respective transcriptional start sites within the narKGHJI and hmp promoter are shown. (B) Mutations introduced within (M2, −61A→G, −62A→C, and −63T→G) and outside (M1, −68C→A, −69C→A, and −70G→A; M3, −55G→T, −56G→T, and −57C→T) the proposed binding site within the narKGHJI promoter are shown below the wild-type (WT) sequence. (C) EMSAs using the wild type and M1, M2, and M3 fragments and purified ArnR-His6 at a molar excess of 0 (minus sign) or 48 (plus sign). (D) DNase I footprinting analysis of ArnR binding to the narK promoter. The IRD-labeled 263-bp narK promoter fragment (2 nM) was incubated with increasing concentrations of ArnR. Lane 1, no protein; lane 2, 60 nM; lane 3, 120 nM; lane 4, 240 nM. The mixture was subjected to partial digestion by DNase I. The sequence protected by ArnR containing the proposed ARS (black box) is shown on the right.
The specific characteristics of the binding of ArnR to the narK promoter region were further assessed by introducing site mutations as follows: 30-bp DNA fragments that bore selected mutations either within the ARS sequence (M2, −61A→G, −62A→C, and −63T→G) or outside of it (M1, −68C→A, −69C→A, and −70G→A; M3, −55G→T, −56G→T, and −57C→T) were generated (Fig. 4B). These mutant ARSs were subjected to EMSAs. As shown in Fig. 4C, ArnR showed specific binding to fragments containing wild-type ARS (WT) and to fragments with mutations outside ARS (M1 and M3), whereas the binding was almost annihilated by mutations within ARS (M2).
DNase I footprinting analysis of the narK promoter region was performed in order to confirm the ARS proposed by EMSAs. As shown in Fig. 4D, the region at positions −68 to −42 relative to the narK transcriptional start site was protected from DNase I cleavage by ArnR. As predicted, the proposed ARS was found within the DNase I-protected region.
The ArnR binding site is involved in the regulation of the expression of the narKGHJI operon and the arnR gene in vivo.
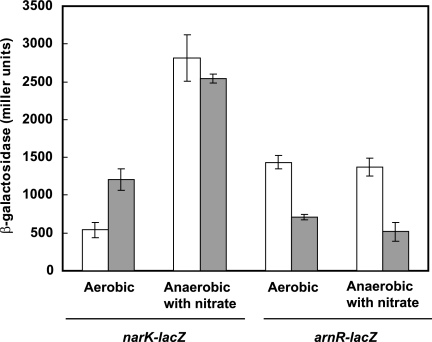
To examine whether ARS is indeed involved in the regulation of the expression of the narKGHJI operon and the arnR gene in vivo, ARS-deleted promoter regions of narKGHJI and arnR were examined for their promoter activities using a lacZ reporter system. ARS-deleted narK- and arnR-lacZ reporter gene fusions were expressed in wild-type C. glutamicum R. These recombinant strains expressing those two fusion proteins were grown in BT minimal medium under aerobic and anaerobic conditions in the presence of nitrate. As shown in Fig. 5, under aerobic conditions, the strain carrying pCRB605, thus lacking ARS from the narKGHJI promoter, exhibited approximately a 2.2-fold-higher β-galactosidase activity than did the strain harboring pCRB603 (intact narK-lacZ fusion). In contrast, both strains showed equally up-regulated levels of β-galactosidase activity under anaerobic conditions. The induced levels were lower than those of the narK and narG genes by QRT-PCR analysis (Table 2), which may be due to the titration of trans-acting regulatory proteins in the promoter assay using multicopy lacZ fusions. The alteration of narKGHJI promoter activity by the deletion of ARS indicates that ARS is responsible for the repression of expression of the narKGHJI operon under aerobic conditions. On the other hand, under both aerobic and anaerobic conditions, wild-type cells harboring pCRB606 lacking ARS from the arnR promoter showed β-galactosidase activities that were decreased by approximately 50% relative to that of the strain carrying pCRB604 (intact arnR-lacZ fusion). This result is consistent with that observed for ΔarnR cells carrying pCRB604 (Fig. 2).
FIG. 5.
Effect of deletion of the deduced ARS on promoter activities of narK and arnR. β-Galactosidase activities (Miller units) of wild-type C. glutamicum strain R harboring various promoter-lacZ fusion plasmids were assessed under conditions of aerobic or anaerobic growth with 30 mM nitrate. White bars, strain harboring an intact narK-lacZ or arnR-lacZ fusion plasmid, pCRB603 or pCRB604, respectively; gray bars, strain harboring an ARS-deleted narK-lacZ or arnR-lacZ fusion plasmid, pCRB605 or pCRB606, respectively. The means ± standard deviations (error bars) of three independent experiments are shown.
C. glutamicum ArnR likely contains an oxygen-labile Fe-S cluster.
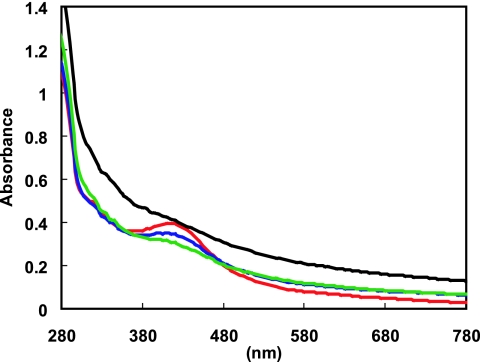
Anoxically purified ArnR protein shows a brownish color, suggesting the presence of a light-absorbing cofactor. UV-visible absorption spectra (280 to 780 nm) of ArnR revealed a shoulder at 320 nm and a broad peak at 420 nm, in addition to the protein absorption maximum at 280 nm (Fig. 6). This profile strikingly resembles that of typical UV-visible absorption Fe-S cluster-containing proteins (13, 31). Moreover, the peak at 420 nm gradually disappeared following 3 h of exposure to air, implying that ArnR possesses an oxygen-sensing cofactor as previously observed with many Fe-S proteins.
FIG. 6.
UV-visible absorption spectrum of anoxically purified ArnR before (red line) and after 1 h (blue line), 2 h (green line), and 3 h (black line) of exposure to air are indicated.
DISCUSSION
We previously reported that the anaerobic growth of C. glutamicum in the presence of nitrate is enabled by the narKGHJI operon, whose expression is regulated by anaerobiosis and nitrate (28). In this study, we identified a gene, arnR, the product of which is a transcriptional regulator that directly represses the expressions of the narKGHJI operon and the hmp gene under aerobic growth conditions. The arnR gene itself was shown to be positively regulated by its own product.
The C. glutamicum ArnR regulator identified in this study is structurally distinct from any Fnr-type regulators that have been shown to this date to activate the expression of nitrate reductase genes in response to anaerobiosis in other bacteria, e.g., E. coli and B. subtilis (4, 25, 32). Fnr-type regulators usually have a carboxyl-terminal DNA-binding domain and three amino-terminally-located cysteine residues together with a central cysteine residue (17), although B. subtilis Fnr exclusively possesses three cysteine residues at the carboxyl terminus that are involved in Fe-S cluster formation (31). Conversely, C. glutamicum ArnR has a DNA-binding domain and three cysteine residues (Cys179, Cys193, and Cys223) at the amino and carboxyl termini, respectively. Our results from gel shift, DNase I footprinting, and lacZ reporter assays revealed the structure of the ArnR binding site, which consists of the 10-bp sequence TA(A/T)TTAA(A/T)TA. This binding site is remarkably distinct from that of the canonical E. coli Fnr box (TTGAT-N4-ATCAA) (37) and from that of the putative canonical B. subtilis Fnr-binding site (TGTGA-N6-TCACA) (32).
The repressor role that ArnR plays in the expression of the narKGHJI operon under aerobic conditions via directly binding to the defined site in the promoter region was confirmed by a narK-lacZ reporter assay, where the deletion of the ArnR binding site upstream of the narK promoter results in increased promoter activity (Fig. 5). In contrast, under anaerobic growth conditions with nitrate, neither the deletion of the arnR coding region nor the deletion of the ArnR binding site from the narK promoter has any effect on the level of expression of the narKGHJI operon, indicating that ArnR plays an important role under aerobic conditions rather than under anaerobic conditions. The aerobic repression of the narKGHJI operon by C. glutamicum ArnR contrasts with the anaerobic activation of the narK gene and the narGHJI operon by E. coli Fnr and B. subtilis Fnr (4, 8, 32). To our knowledge, the aerobic repression of the nitrate reductase operon has not yet been observed in any other bacterial nitrate respiration systems (49).
The expression level of ArnR is constitutive irrespective of the presence of oxygen (Fig. 2), suggesting that ArnR could become functional as a consequence of a direct interaction with oxygen or another biochemical signal. ArnR possesses an oxygen-sensing cofactor such as an Fe-S cluster (Fig. 6). The cofactor might be coordinated by three cysteine residues of ArnR and involved in the control of the ArnR activity in response to the oxygen/redox state in the cell. A variety of transcriptional regulators are known to have heme, flavin, and an Fe-S cluster as a cofactor. Many of these transcriptional regulators are involved in the oxygen/redox control of target gene expression via a variety of mechanisms (2, 14). Further study will be required to clarify the oxygen-sensing mechanism of ArnR.
In addition to the narKGHJI operon, the arnR and hmp genes were identified as being additional targets of ArnR. Notably, the arnR gene was shown to be positively regulated by its own product irrespective of the presence of oxygen (Fig. 2). This autoinduction system possibly contributes to the maintenance of the constant and optimal expression level of ArnR. On the other hand, ArnR strongly represses the expression of the flavohemoglobin-encoding hmp gene under aerobic conditions (Table 3), the expression of which is significantly induced during anaerobic growth using nitrate (see above). No homologues to NsrR and NorR, which are involved in the regulation of hmp expression in the other bacteria studied (1, 3, 23, 33), are found on the C. glutamicum genome. The physiological function of flavohemoglobin in protecting E. coli from nitric oxide has been firmly established (19, 29, 30). Flavohemoglobin detoxifies nitric oxide by anaerobically reducing it to N2O or by aerobically oxidizing it to nitrate. In B. subtilis, hmp has an essential role in long-term anaerobic survival only in nitrate-containing medium (22). Although C. glutamicum Hmp is not highly similar to either E. coli Hmp (32% identity) or B. subtilis Hmp (31% identity), C. glutamicum Hmp may play some protective role against possible oxidative stress during anaerobic growth with nitrate. Nonetheless, the role that flavohemoglobin plays in the physiology of C. glutamicum needs to be elucidated further.
In the present study, ArnR was identified as being a novel transcriptional regulator that is involved in regulating anaerobic nitrate reduction processes in C. glutamicum. ArnR homologues are found on the genome in other Corynebacterium, Mycobacterium, and Nocardioides species. Similar regulatory systems of nitrate reductase operons are expected to exist in these closely related species, although none of the upstream regions of the nar operons sequenced to date from these species nor the arnR and hmp genes of these bacteria contain any clear ArnR consensus binding site as identified in the present study. On the other hand, the regulation of nitrate reduction in C. glutamicum may be performed by other factors that would act in an Fnr-like manner because the expression of the narKGHJI operon is indeed induced in arnR-deficient cells by an anaerobic shift in the presence of nitrate (Table 2). Notably, previous reports classified three gene products from C. glutamicum R, namely, CgR0133, GlxR (15, 18), and CgR1254, into the Crp/Fnr family of regulators (6). At present, the mechanism of nitrate-dependent regulation of the narKGHJI operon remains unclear, and thus, we will need to precisely examine the degree of involvement of these genes in the regulation of gene expression in response to anaerobiosis. In addition, we will focus our future work on the search for additional regulators of genes involved in anaerobic induction and nitrate sensing.
Supplementary Material
Acknowledgments
This work was financially supported in part by the New Energy and Industrial Technology Development Organization, Japan.
Footnotes
Published ahead of print on 22 February 2008.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Arai, H., M. Hayashi, A. Kuroi, M. Ishii, and Y. Igarashi. 2005. Transcriptional regulation of the flavohemoglobin gene for aerobic nitric oxide detoxification by the second nitric oxide-responsive regulator of Pseudomonas aeruginosa. J. Bacteriol. 1873960-3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer, C. E., S. Elsen, and T. H. Bird. 1999. Mechanisms for redox control of gene expression. Annu. Rev. Microbiol. 53495-523. [DOI] [PubMed] [Google Scholar]
- 3.Bodenmiller, D. M., and S. Spiro. 2006. The yjeB (nsrR) gene of Escherichia coli encodes a nitric oxide-sensitive transcriptional regulator. J. Bacteriol. 188874-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonnefoy, V., and J. A. Demoss. 1994. Nitrate reductases in Escherichia coli. Antonie van Leeuwenhoek 6647-56. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 6.Brune, I., K. Brinkrolf, J. Kalinowski, A. Püehler, and A. Tauch. 2005. The individual and common repertoire of DNA-binding transcriptional regulators of Corynebacterium glutamicum, Corynebacterium efficiens, Corynebacterium diphtheriae and Corynebacterium jeikeium deduced from the complete genome sequences. BMC Genomics 686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clegg, S., F. Yu, L. Griffiths, and J. A. Cole. 2002. The roles of the polytopic membrane proteins NarK, NarU and NirC in Escherichia coli K-12: two nitrate and three nitrite transporters. Mol. Microbiol. 44143-155. [DOI] [PubMed] [Google Scholar]
- 8.Cruz Ramos, H., L. Boursier, I. Moszer, F. Kunst, A. Danchin, and P. Glaser. 1995. Anaerobic transcription activation in Bacillus subtilis: identification of distinct FNR-dependent and -independent regulatory mechanisms. EMBO J. 145984-5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green, J., B. Bennett, P. Jordan, E. T. Ralph, A. J. Thomson, and J. R. Guest. 1996. Reconstitution of the [4Fe-4S] cluster in FNR and demonstration of the aerobic-anaerobic transcription switch in vitro. Biochem. J. 316887-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gunsalus, R. P., and S. J. Park. 1994. Aerobic-anaerobic gene regulation in Escherichia coli: control by the ArcAB and Fnr regulons. Res. Microbiol. 145437-450. [DOI] [PubMed] [Google Scholar]
- 11.Inui, M., H. Kawaguchi, S. Murakami, A. A. Vertès, and H. Yukawa. 2004. Metabolic engineering of Corynebacterium glutamicum for fuel ethanol production under oxygen-deprivation conditions. J. Mol. Microbiol. Biotechnol. 8243-254. [DOI] [PubMed] [Google Scholar]
- 12.Inui, M., M. Suda, S. Okino, H. Nonaka, L. G. Puskas, A. A. Vertès, and H. Yukawa. 2007. Transcriptional profiling of Corynebacterium glutamicum metabolism during organic acid production under oxygen deprivation conditions. Microbiology 1532491-2504. [DOI] [PubMed] [Google Scholar]
- 13.Khoroshilova, N., C. Popescu, E. Münck, H. Beinert, and P. J. Kiley. 1997. Iron-sulfur cluster disassembly in the FNR protein of Escherichia coli by O2: [4Fe-4S] to [2Fe-2S] conversion with loss of biological activity. Proc. Natl. Acad. Sci. USA 946087-6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiley, P. J., and H. Beinert. 2003. The role of Fe-S proteins in sensing and regulation in bacteria. Curr. Opin. Microbiol. 6181-185. [DOI] [PubMed] [Google Scholar]
- 15.Kim, H. J., T. H. Kim, Y. Kim, and H. S. Lee. 2004. Identification and characterization of glxR, a gene involved in regulation of glyoxylate bypass in Corynebacterium glutamicum. J. Bacteriol. 1863453-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kinoshita, S. 1985. Glutamic acid bacteria, p. 115-146. In A. L. Demain and N. A. Solomon (ed.), Biology of industrial microorganisms. Benjamin Cummings, London, United Kingdom.
- 17.Körner, H., H. J. Sofia, and W. G. Zumft. 2003. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 27559-592. [DOI] [PubMed] [Google Scholar]
- 18.Letek, M., N. Valbuena, A. Ramos, E. Ordóñez, J. A. Gil, and L. M. Mateos. 2006. Characterization and use of catabolite-repressed promoters from gluconate genes in Corynebacterium glutamicum. J. Bacteriol. 188409-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Membrillo-Hernández, J., M. D. Coopamah, M. F. Anjum, T. M. Stevanin, A. Kelly, M. N. Hughes, and R. K. Poole. 1999. The flavohemoglobin of Escherichia coli confers resistance to a nitrosating agent, a “nitric oxide releaser,” and paraquat and is essential for transcriptional responses to oxidative stress. J. Biol. Chem. 274748-754. [DOI] [PubMed] [Google Scholar]
- 20.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 21.Moreno-Vivián, C., P. Cabello, M. Martínez-Luque, R. Blasco, and F. Castillo. 1999. Prokaryotic nitrate reduction: molecular properties and functional distinction among bacterial nitrate reductases. J. Bacteriol. 1816573-6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakano, M. M. 2006. Essential role of flavohemoglobin in long-term anaerobic survival of Bacillus subtilis. J. Bacteriol. 1886415-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakano, M. M., H. Geng, S. Nakano, and K. Kobayashi. 2006. The nitric oxide-responsive regulator NsrR controls ResDE-dependent gene expression. J. Bacteriol. 1885878-5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakano, M. M., Y. Zhu, M. Lacelle, X. Zhang, and F. M. Hulett. 2000. Interaction of ResD with regulatory regions of anaerobically induced genes in Bacillus subtilis. Mol. Microbiol. 371198-1207. [DOI] [PubMed] [Google Scholar]
- 25.Nakano, M. M., and P. Zuber. 1998. Anaerobic growth of a “strict aerobe” (Bacillus subtilis). Annu. Rev. Microbiol. 52165-190. [DOI] [PubMed] [Google Scholar]
- 26.Nakano, M. M., P. Zuber, P. Glaser, A. Danchin, and F. M. Hulett. 1996. Two-component regulatory proteins ResD-ResE are required for transcriptional activation of fnr upon oxygen limitation in Bacillus subtilis. J. Bacteriol. 1783796-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakata, K., M. Inui, P. B. Kos, A. A. Vertès, and H. Yukawa. 2003. Vectors for the genetics engineering of corynebacteria, p. 175-191. In B. C. Saha (ed.), Fermentation biotechnology, ACS Symposium series 862. American Chemical Society, Washington, DC.
- 28.Nishimura, T., A. A. Vertès, Y. Shinoda, M. Inui, and H. Yukawa. 2007. Anaerobic growth of Corynebacterium glutamicum using nitrate as a terminal electron acceptor. Appl. Microbiol. Biotechnol. 75889-897. [DOI] [PubMed] [Google Scholar]
- 29.Poole, R. K., M. F. Anjum, J. Membrillo-Hernández, S. O. Kim, M. N. Hughes, and V. Stewart. 1996. Nitric oxide, nitrite, and Fnr regulation of hmp (flavohemoglobin) gene expression in Escherichia coli K-12. J. Bacteriol. 1785487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poole, R. K., and M. N. Hughes. 2000. New functions for the ancient globin family: bacterial responses to nitric oxide and nitrosative stress. Mol. Microbiol. 36775-783. [DOI] [PubMed] [Google Scholar]
- 31.Reents, H., I. Gruner, U. Harmening, L. H. Böttger, G. Layer, P. Heathcote, A. X. Trautwein, D. Jahn, and E. Härtig. 2006. Bacillus subtilis Fnr senses oxygen via a [4Fe-4S] cluster coordinated by three cysteine residues without change in the oligomeric state. Mol. Microbiol. 601432-1445. [DOI] [PubMed] [Google Scholar]
- 32.Reents, H., R. Münch, T. Dammeyer, D. Jahn, and E. Härtig. 2006. The Fnr regulon of Bacillus subtilis. J. Bacteriol. 1881103-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodionov, D. A., I. L. Dubchak, A. P. Arkin, E. J. Alm, and M. S. Gelfand. 2005. Dissimilatory metabolism of nitrogen oxides in bacteria: comparative reconstruction of transcriptional networks. PLoS Comput. Biol. 1e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 35.Schreiber, K., R. Krieger, B. Benkert, M. Eschbach, H. Arai, M. Schobert, and D. Jahn. 2007. The anaerobic regulatory network required for Pseudomonas aeruginosa nitrate respiration. J. Bacteriol. 1894310-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spiro, S. 1994. The FNR family of transcriptional regulators. Antonie van Leeuwenhoek 6623-36. [DOI] [PubMed] [Google Scholar]
- 37.Spiro, S., and J. R. Guest. 1990. FNR and its role in oxygen-regulated gene expression in Escherichia coli. FEMS Microbiol. Rev. 6399-428. [DOI] [PubMed] [Google Scholar]
- 38.Spiro, S., and J. R. Guest. 1987. Regulation and over-expression of the fnr gene of Escherichia coli. J. Gen. Microbiol. 1333279-3288. [DOI] [PubMed] [Google Scholar]
- 39.Stewart, V. 1993. Nitrate regulation of anaerobic respiratory gene expression in Escherichia coli. Mol. Microbiol. 9425-434. [DOI] [PubMed] [Google Scholar]
- 40.Stewart, V. 1994. Regulation of nitrate and nitrite reductase synthesis in enterobacteria. Antonie van Leeuwenhoek 6637-45. [DOI] [PubMed] [Google Scholar]
- 41.Sun, G., E. Sharkova, R. Chesnut, S. Birkey, M. F. Duggan, A. Sorokin, P. Pujic, S. D. Ehrlich, and F. M. Hulett. 1996. Regulators of aerobic and anaerobic respiration in Bacillus subtilis. J. Bacteriol. 1781374-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takeno, S., J. Ohnishi, T. Komatsu, T. Masaki, K. Sen, and M. Ikeda. 2007. Anaerobic growth and potential for amino acid production by nitrate respiration in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 751173-1182. [DOI] [PubMed] [Google Scholar]
- 43.Terasawa, M., and H. Yukawa. 1993. Industrial production of biochemicals by native immobilization, p. 37-52. In A. Tanaka, T. Tosa, and T. Kobayashi (ed.), Industrial application of immobilized biocatalysts. Dekker, New York, NY. [PubMed]
- 44.Unden, G., S. Achebach, G. Holighaus, Q. H. Tran, B. Wackwitz, and Y. Zeuner. 2002. Control of FNR function of Escherichia coli by O2 and reducing conditions. J. Mol. Microbiol. Biotechnol. 4263-268. [PubMed] [Google Scholar]
- 45.Vertès, A. A., M. Inui, M. Kobayashi, Y. Kurusu, and H. Yukawa. 1993. Presence of mrr- and mcr-like restriction systems in coryneform bacteria. Res. Microbiol. 144181-185. [DOI] [PubMed] [Google Scholar]
- 46.Wennerhold, J., A. Krug, and M. Bott. 2005. The AraC-type regulator RipA represses aconitase and other iron proteins from Corynebacterium under iron limitation and is itself repressed by DtxR. J. Biol. Chem. 28040500-40508. [DOI] [PubMed] [Google Scholar]
- 47.Ye, R. W., W. Tao, L. Bedzyk, T. Young, M. Chen, and L. Li. 2000. Global gene expression profiles of Bacillus subtilis grown under anaerobic conditions. J. Bacteriol. 1824458-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yukawa, H., C. A. Omumasaba, H. Nonaka, P. Kos, N. Okai, N. Suzuki, M. Suda, Y. Tsuge, J. Watanabe, Y. Ikeda, A. A. Vertès, and M. Inui. 2007. Comparative analysis of the Corynebacterium glutamicum group and complete genome sequence of strain R. Microbiology 1531042-1058. [DOI] [PubMed] [Google Scholar]
- 49.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.