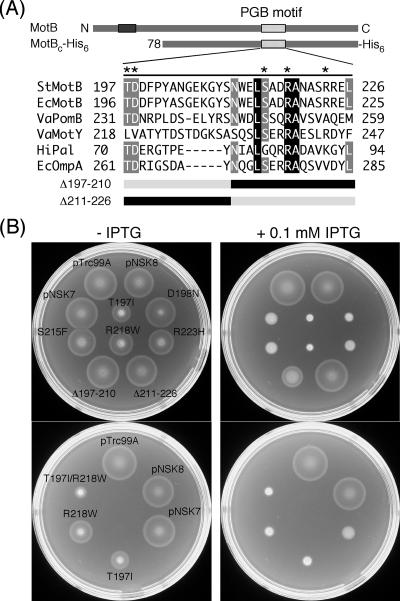
FIG. 2.
Dominance properties of various mutant variants of MotBC-His6. (A) Multiple-sequence alignment of bacterial proteins containing a PGB motif and mutations generated in MotBC-His6. Sequences aligned are as follows: StMotB, Salmonella enterica serovar Typhimurium MotB; EcMotB, Escherichia coli MotB; VaPomB, Vibrio alginolyticus PomB; VaMotY, Vibrio alginolyticus MotY; HiPal, Haemophilus influenzae Pal; EcOmpA, Escherichia coli OmpA. The multiple-sequence alignment was done by the ClustalW software (42). Asterisks indicate residues mutated in this study. Residues shown in the black (or gray) box with white letters are completely conserved (or well conserved) among these six proteins. The point mutations are those originally reported by Blair et al. (6) to produce a dominant-negative Mot− phenotype in E. coli. Δ197-210 and Δ211-226 are deletion mutants that lack residues 197 to 210 and 211 to 226, respectively. (B) Swarming motility assay of wild-type strain SJW1103 transformed with plasmids pTrc99A (vector control), pNSK8 (MotBC-His6), pNSK7 (PelBL-MotBC-His6), T197I [PelBL-MotBC(T197I)-His6], D198N [PelBL-MotBC(D198N)-His6], S215F [PelBL-MotBC(S215F)-His6], R218W [PelBL-MotBC(R218W)-His6], R223H [PelBL-MotBC(R223H)-His6], Δ197-210 [PelBL-MotBC(Δ197-210)-His6), Δ211-226 [PelBL-MotBC(Δ211-226)-His6), and T197I/R218W [PelBL-MotBC(T197I/R218W)-His6]. Cells were inoculated onto the same positions of soft-agar plates with or without 0.1 mM IPTG. The strain names are indicated only on the left plate. Plates were incubated at 30°C for 6 h.