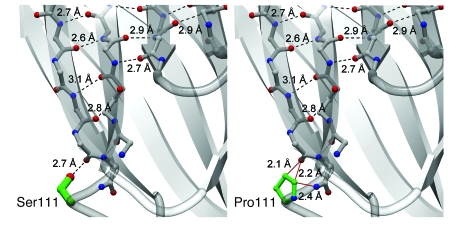
Figure 9. Effects of proline substitution at residue 111 of cPLA2α.
Comparison of the wild-type cPLA2α structure (left) with a model of the p.[S111P] mutation (right). Backbone carbon atoms are shown in gray, oxygen atoms and hydroxy groups in red, nitrogen atoms in blue, and side-chain carbon atoms of the 111 position in green. Hydrogen-bonding interactions are shown in dashed black lines and sterically unfavorable close distances are shown in solid red lines. A hydrogen-bonding interaction has an ideal distance of 2.7–3.0 υ between oxygen and nitrogen-hydrogen–bond donors and acceptors and an ideal distance of 3.1–3.2 υ for sulfur to nitrogen-hydrogen–bond donors and acceptors. Nonbonded distances smaller than 2.5 υ between nitrogen and oxygen-hydrogen–bonding donors and acceptors are sterically and energetically unfavorable and are generally not observed in nature. Distances longer than 3.5 υ do not contribute favorable energy toward folding stabilization. Modeling of proline in position 111 reveals that 1 ideal hydrogen-bonding interaction of 2.7 υ between the p.[S111] side-chain hydroxyl moiety and the backbone carbonyl group of p.[T108] has been replaced by 3 sterically unfavorable interactions (<2.5 υ). Two of these unfavorable interactions lie between the hydrophobic proline Cγ and Cδ atoms and the p.[T108] carbonyl, while 1 unfavorable interaction is between the proline Cδ and the amide nitrogen of p.[S111]. The p.[S111P] mutation is predicted to cause unfavorable interactions (red lines) in the absence of a structural rearrangement of the protein. The predicted decrease in stability of the adjacent β-strands likely affects the entire Ca2+-binding domain.