Abstract
Vaccines exert strong selective pressures on pathogens, favouring the spread of antigenic variants. We propose a simple mathematical model to investigate the dynamics of a novel pathogenic strain that emerges in a population where a previous strain is maintained at low endemic level by a vaccine. We compare three methods to assess the ability of the novel strain to invade and persist: algebraic rate of invasion; deterministic dynamics; and stochastic dynamics. These three techniques provide complementary predictions on the fate of the system. In particular, we emphasize the importance of stochastic simulations, which account for the possibility of extinctions of either strain. More specifically, our model suggests that the probability of persistence of an invasive strain (i) can be minimized for intermediate levels of vaccine cross-protection (i.e. immune protection against the novel strain) and (ii) is lower if cross-immunity acts through a reduced infectious period rather than through reduced susceptibility.
Keywords: cross-immunity, antigenic diversity, Bordetella pertussis
1. Introduction
Antigenic variation of pathogens is a well-known phenomenon, but it still poses a threat to the control of infectious diseases. The prospect of efficient vaccines against HIV, dengue or malaria is hampered by the existing antigenic diversity. Vaccines have the potential to impose strong selective pressure on pathogens, allowing the spread of escape mutants. For reasons mostly unknown, some infections have been effectively controlled by mass vaccination without evolving escape mutants. This is the case for most childhood diseases (measles, mumps, rubella, diphtheria, poliomyelitis, etc.), as well as smallpox, the only example of global eradication to date. But the success of other immunization campaigns has been challenged by the recent outbreaks caused by non-vaccine strains. A growing body of evidence is pointing to vaccine-driven antigenic evasion in at least four diseases across the world: (i) hepatitis B virus, in Italy (Carman et al. 1990), Taiwan (Hsu et al. 1999), The Gambia (Karthigesu et al. 1999), Singapore (Chen & Oon 2000) and Turkey (Türkyilmaz et al. 2006), (ii) Bordetella pertussis in the Netherlands (Mooi et al. 1998), Italy (Mastrantonio et al. 1999), the USA (Cassiday et al. 2000), France (Weber et al. 2001), Poland (Gzyl et al. 2002), Canada (Peppler et al. 2003), the UK (Packard et al. 2004), Finland (Elomaa et al. 2005), Argentina (Fingermann et al. 2006) and Taiwan (Lin et al. 2006), (iii) Streptococcus pneumoniae, in The Gambia (Obaro et al. 1996), the USA (Brook et al. 2006; Flannery et al. 2006) and South Africa (Mbelle et al. 1999), and (iv) Haemophilus influenzae (non-type b) meningitis, in Alaska (Perdue et al. 2000), Brazil (de Almeida et al. 2005), and Canada (Tsang et al. 2006).
Mathematical models have been developed to help understand the factors that can promote the emergence of escape mutants in the presence of a vaccine. However, whereas multiple-strain models without vaccine have flourished for the last 10 years (Andreasen et al. 1997; Gupta et al. 1998; Ferguson et al. 1999; Dawes & Gog 2002; Gog & Grenfell 2002; Gomes et al. 2002; Grenfell et al. 2004), models for vaccine evasion have seen limited development since a series of seminal articles published around 10 years ago (McLean 1995; Gupta et al. 1997; Lipsitch 1997; White et al. 1998). They considered at least two strains with different basic reproductive ratios (R0) and a vaccine that targets the best competitor(s). They derived conditions (vaccine coverage, cross-protection, etc.) that allow less fit strains to spread, with or without excluding competitors. Since then, similar models have been applied to some of the diseases mentioned previously: hepatitis B (Wilson et al. 1999) and pneumococcal disease (Zhang et al. 2004), but also human papillomavirus (Elbasha & Galvani 2005) ahead of vaccine implementation.
All those models rely on deterministic dynamics and base their predictions on the following framework: if the initial growth rate of strain x in the presence of strain y is positive, but the opposite does not hold, then only strain x will remain at equilibrium. Alternatively, if the strains can invade each other, then the usual outcome is stable coexistence (or possibly oscillations when more than two strains are considered). However, including chance (i.e. demographic stochasticity) into models is likely to alter those simple predictions. Stochastic models for infectious disease dynamics have been studied in great details (Bartlett 1953; Keeling & Grenfell 1997; Mollison 1977; van Herwaarden 1997; Andersson & Britton 2000; Grenfell et al. 2001; Nåsell 2002), showing, in particular, that even ‘fit’ pathogens can go extinct in the turmoil of epidemic dynamics. Recent models have started to explore the interactions between strain diversity and epidemiological dynamics in stochastic frameworks (Gog et al. 2003; Abu-Raddad & Ferguson 2004; Kirupaharan & Allen 2004; Stollenwerk et al. 2004; Restif & Grenfell 2006), but vaccination has not been taken into account yet.
In this paper, we study immune evasion as a dynamic process, in which an antigenic mutant may invade but not necessarily persist. Practically, in a population where an endemic pathogenic strain is maintained at a low incidence by a vaccine, we introduce a mutant strain against which vaccination confers limited protection. In contrast with the studies cited previously, we assume that the novel strain has the same basic reproductive ratio as the endemic strain. Therefore, in a deterministic framework, the mutant is able to invade with or without vaccine, and the two strains are expected to coexist. However, in a stochastic framework, we show that invasion can be rapidly followed by the extinction of either strain and that the probability of persistence depends on the qualitative and quantitative characteristics of the cross-immunity conferred by the vaccine against the novel strain.
Following on our previous study (Restif & Grenfell 2006), in which there was no vaccination, we choose pertussis as a topical example to illustrate our point. However, the same method can be applied to other acute infectious diseases. By combining different modelling approaches, we aim to provide more complete predictions on the shorter- and longer-term effects of vaccination on both antigenic variation and epidemic dynamics.
2. Methods
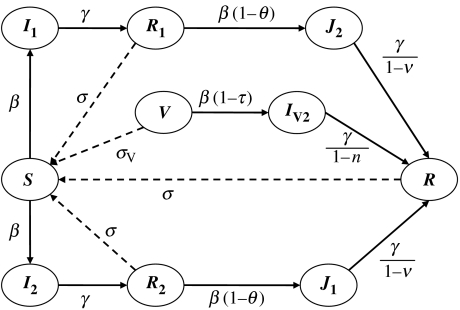
The model (figure 1) is derived from a classical susceptible–infected–recovered (SIR) model with a few additions. First, we consider two strains of the pathogen which interact through cross-infection. In the following, strain 1 is initially endemic, whereas strain 2 is a new strain introduced into the population. Second, a vaccine is used to immunize a proportion p of all newborns, conferring full protection against strain 1, but partial protection only against strain 2. Third, cross-protection (conferred by the vaccine or the primary infection) is modelled as either reduced infection rate or shorter infectious period (table 1); for each of the four cross-protection parameters, a value of zero represents no change, and cross-immunity increases as the value approaches 1, which represents total protection. Lastly, immunity wanes at a constant rate (σ for natural infection, σV for vaccination).
Figure 1.
Compartmental sketch of the model used in this study. Births and deaths at rate μ per capita are not shown, nor vaccine uptake p.
Table 1.
The four types of cross-protection.
| effect | source | |
|---|---|---|
| natural infection | vaccination | |
| on infection rate β | β(1−θ) | β(1−τ) |
| on infectious period 1/γ | (1−ν)/γ | (1−η)/γ |
In order to keep the model reasonably simple, we make a number of simplifying assumptions, which could be easily modified in line with relevant empirical data. Here, these parsimonious assumptions reflect the lack of information currently available on pertussis. In particular, both strains are assumed to have the same infection rate β, infectious period 1/γ and immune period 1/σ, and all infected individuals are equally infectious. In addition, births and deaths occur at the same rate μ, so that the population size remains constant in the deterministic model.
We successively used three techniques to explore the dynamics of this system. First, we analysed the initial invasion rate of the novel strain, using a deterministic version of the model, as described by the following system of ordinary differential equations (see table 2 for a description of symbols):
| (2.1) |
Table 2.
Symbols used.
| S | proportion susceptible to both strains |
| Ii | proportion in primary infection with strain i={1,2} |
| Ri | proportion immune to strain i only |
| Ji | proportion in secondary infection with strain i={1,2} |
| R | proportion immune to both strains |
| V | proportion vaccinated and still immune to strain 1 |
| IV2 | proportion vaccinated and infected by strain 2 |
| β | rate of transmission |
| γ | rate of recovery |
| μ | birth and death rate |
| p | vaccine coverage (proportion of newborns vaccinated) |
| σ | rate of loss of natural immunity |
| σV | rate of loss of vaccine immunity |
| θ | cross-protection conferred by primary infection (reduction in susceptibility) |
| ν | cross-protection conferred by primary infection (reduction in infectious period) |
| τ | cross-protection conferred by vaccination (reduction in susceptibility) |
| η | cross-protection conferred by vaccination (reduction in infectious period) |
| R0 | basic reproductive ratio of the pathogen: R0=β/(γ+μ) |
Second, we ran deterministic simulations by numerically integrating the above system, starting from endemic equilibrium of strain 1 with vaccine coverage p and introducing a fraction of 10−6 infected with the novel strain 2. We recorded the depth of the post-epidemic trough, i.e. the minimum fractions infected with strain 1 or 2 following introduction.
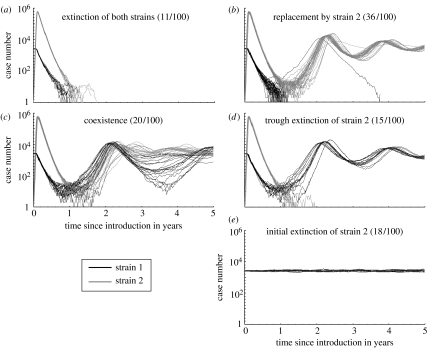
Third, we implemented a stochastic version of the model using a Gillespie-type algorithm (Bartlett 1953; Gillespie 1977; Gibson & Bruck 2000), as described by Restif & Grenfell (2006). Simulations were run with an initial population size of one million, starting from the (deterministic) endemic equilibrium for strain 1 and introducing one infected individual with strain 2 at time t=0. For each set of parameter values, we ran a series of 1000 simulations for 10 years, during which we recorded all infections, as well as extinctions of either strain. Simulations were then classified based on the events of extinction within 10 years of introduction: extinction of both strains; initial extinction of strain 2 (i.e. without causing an outbreak); trough extinction of strain 2 only; replacement by strain 2 (i.e. only strain 1 went extinct after the outbreak); or coexistence (no extinction). Because all recorded extinctions occurred within 5 years of the introduction, we are confident that the outcome after 10 years is reliable as to the long-term persistence of strains in the population. However, that may not be the case in small populations, as endemic levels would be much lower and more prone to fade-outs.
Overall, we focused on the effect of vaccine parameters (coverage and cross-protection). Numerical values were inspired by pertussis, with a high basic reproductive ratio R0=17, an average infectious period of three weeks and average immune periods of several years (Anderson & May 1991; Esposito et al. 2001; Broutin et al. 2004).
3. Results
3.1 Invasion analysis
We first consider the situation where strain 1 is endemic in the presence of the vaccine and strain 2 is absent. The system admits an equilibrium given by
| (3.1) |
where R0=β/(μ+γ) is the pathogen's basic reproductive ratio. It can be shown (Moghadas & Gumel 2003) that the equilibrium is stable if the two following conditions are met:
| (3.2) |
Hence, elimination of the disease by vaccination is feasible only if σV<μ/(R0−1). In other words, the average immune period of the vaccine (1/σV) must be at least (R0−1) times the average lifespan (1/μ), which is far from being the case for pertussis vaccines (Anderson & May 1991; Esposito et al. 2001; Broutin et al. 2004) and probably many others. In the following, we will assume that
| (3.3) |
so that strain 1 is endemic whatever the vaccine coverage.
In a second step, we consider that strain 2 is introduced into the population at a very low frequency. It can be shown (see appendix A) that, after short transient dynamics, the frequency of strain 2 (I2+J2+IV2) will approximately follow an exponential growth of the form exp(xt), where x can be found numerically by solving the following equation:
| (3.4) |
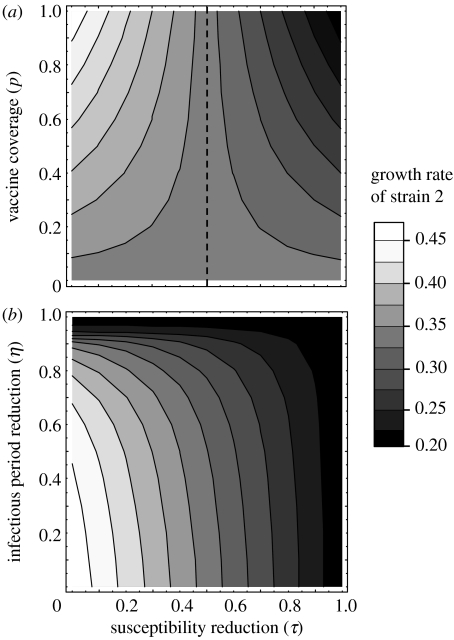
which always admits one positive root, so strain 2 can always spread in the population (unless cross-protection is perfect, i.e. θ=τ=1 or ν=η=1). This growth rate x depends on vaccine coverage and cross-protection, as shown in figure 2. In particular, higher coverage enhances the spread of the novel strain if vaccination is less protective against cross-infection than natural immunity, i.e. τ<θ. Using different models, McLean (1995), Lipsitch (1997) and White et al. (1998) previously showed that increasing the coverage of a vaccine with limited cross-protection can favour the invasion of a novel strain, even if the latter has a lower R0 than the endemic strain.
Figure 2.
The greyscale represents the initial growth rate of strain 2 (obtained by solving equation (3.4) for x), as a function of cross-protection on susceptibility τ on the horizontal axis, and (a) vaccine coverage p or (b) reduction in infectious period η on the vertical axis. In (a), the left-hand side of the vertical dashed line corresponds to a vaccine conferring narrow antigenic protection, τ<θ. Parameter values: R0=17; θ=0.5; average infectious period 1/γ=21 days; average immune periods 1/σ=1/σV=20 years; average lifespan 1/μ=50 years; (a) no reduction in infectious period through cross-immunity (ν=η=0); and (b) vaccine coverage p=1 and ν=0.5.
Figure 2b illustrates the asymmetry between the two forms of vaccine cross-protection considered in the model. Indeed, a vaccine which reduces susceptibility to a novel strain without affecting its infectious period may be less permissive than a vaccine that shortens the infectious period of the novel strain without affecting susceptibility. The same argument can be made from the viewpoint of the mutant strain: there appears to be an advantage to evading vaccine protection on susceptibility (low τ but high η) over evading protection on infectious period (low η but high τ).
3.2 Deterministic dynamics
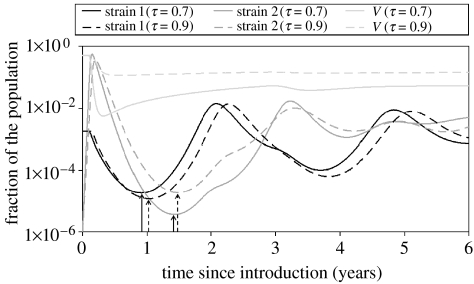
Numerical simulations confirmed that the deterministic system always converges towards stable coexistence of the two strains, under the conditions stated previously. However, as argued in our previous paper (Restif & Grenfell 2006), the depth of the post-epidemic troughs, i.e. the minimal incidence reached by each strain after the initial outbreak (figure 3), suggests that, in a finite population, one strain or even both might go extinct. Actual frequencies of extinction will be studied in §3.3 using a stochastic framework. Here, we record the depth of troughs and illustrate the effects of vaccine parameters on epidemic dynamics, beyond simple invasion analyses.
Figure 3.
Deterministic trajectories, obtained by the numerical integration of equations (2.1), following the introduction of strain 2 with an incidence of 10−6. Black lines show the incidence (infected fraction of the population) of strain 1, darker grey lines that of strain 2 and lighter grey lines the fraction in the vaccinated compartment V. Solid lines, vaccine cross-protection τ=0.7; dashed lines, τ=0.9. Arrows point at post-epidemic troughs, as recorded in figures 5 and 6. Other parameter values are as in figure 2, except vaccine immune period 1/σV=50 years.
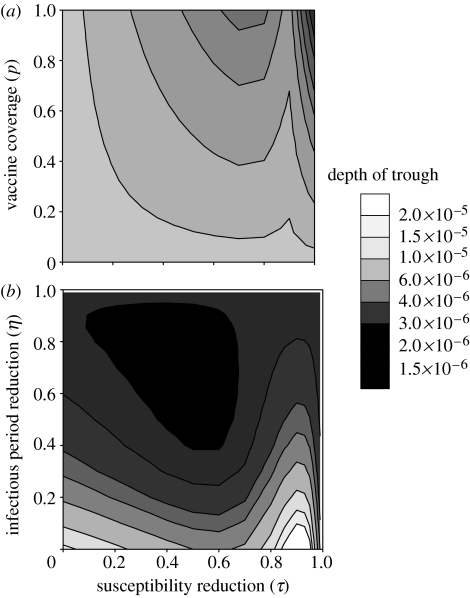
We showed in §3.1 that high vaccination coverage and long-lasting immunity can favour the initial spread of strain 2, if vaccine-induced cross-immunity is weaker than natural cross-immunity (figure 2a). As illustrated in figure 4, the trough patterns are more complex. First, increasing vaccine coverage (figure 4a) always leads to deeper troughs of strain 2, as well as strain 1 (not shown). If vaccine cross-protection is weaker than natural cross-protection (τ<θ), high vaccine coverage has a weaker, but still negative, effect on trough depth of strain 2; this suggests that the initial benefit of invasion conferred by high vaccine coverage can be followed by a higher risk of extinction.
Figure 4.
Trough depth of strain 2 plotted against the reduction in susceptibility through vaccine cross-immunity (τ) on the horizontal and the vertical axes: (a) vaccine coverage p and (b) reduction in infectious period through vaccine cross-immunity η. Other parameter values are as in figure 2, except (b) ν=0.5.
Second, the effect of vaccine cross-protection on trough depth can be either positive or negative. Actually, the minimal incidence of strain 2 reaches its lowest value for intermediate values of τ and η (e.g. τ=0.5 and η=0.6 in figure 4b). This is caused by two antagonistic effects of cross-protection. On the one hand, weak cross-protection enables a strain to rapidly infect individuals immune to the other strain. On the other hand, strong cross-protection prevents the strain from overexploiting that resource. Figure 3 illustrates the latter effect: stronger cross-protection (τ=0.9 compared with τ=0.7) slows down the initial outbreak and prevents a drastic depletion of the vaccinated compartment (V), so that strain 2 is maintained at a higher incidence during the trough. However, this positive effect of cross-protection has its limits; as can be seen in figure 4b, very high values of τ (typically, τ>0.9) result in deeper troughs. Indeed, such levels of cross-protection strongly reduce the infection rate of strain 2 and hinder its bounce after the trough.
Finally, we showed in §3.1 that a vaccine that reduces susceptibility to new strains (i.e. with a high τ and a low η) is less permissive to invasion than the one that reduces their infectious period (high η and low τ). However, the present analysis indicates that the latter type of vaccine can lead an invasive strain to a deeper post-epidemic trough than the former (figure 4b), hence possibly making its extinction more probable.
3.3 Stochastic simulations
Sections 3.1 and 3.2 highlighted discrepancies between the effects of certain parameters of the model (vaccine coverage and strength of cross-protection) on the initial rate of invasion of strain 2 (§3.1) and the depth of the post-epidemic troughs (§3.2). How informative are those two techniques on the efficacy of a particular vaccine against novel strains or on the evolutionary success of a particular mutant?
Stochastic simulations allow us to track strain invasion and extinction, together with epidemiological dynamics. Figure 5 shows typical trajectories following the introduction of strain 2 in a population. Each possible outcome (depending on whether each strain goes extinct or survives) has a specific signature in terms of total observed cases. For example, survival of one strain only after the initial outbreak (figure 5b or d) tends to cause slower and deeper oscillations (of total cases) than the coexistence of both strains (figure 5c). We are planning to investigate in a further study how the particular shape of these dynamics would be affected by other factors, such as seasonal forcing or population structure, previously included in single-strain models for pertussis (Grenfell & Anderson 1989; Rohani et al. 2002).
Figure 5.
Series of 100 stochastic simulations, grouped by outcome (see §2) in (a)–(e), showing the number of individuals infected with strains 1 (black) and 2 (grey). Parameter values: initial population size, 1 million; average life span 1/μ=50 years; R0=17; vaccine coverage p=1; infectious period 21 days; immune periods 1/σ =1/σV=20 years; natural cross-protection θ=0.6; vaccine cross-protection τ=0.2; and no reduction in infectious period ν=η=0.
We then analyse the effects of vaccine parameters on the frequencies of extinctions and the number of cases following an outbreak. How do they compare with earlier predictions based on invasion rates and deterministic dynamics?
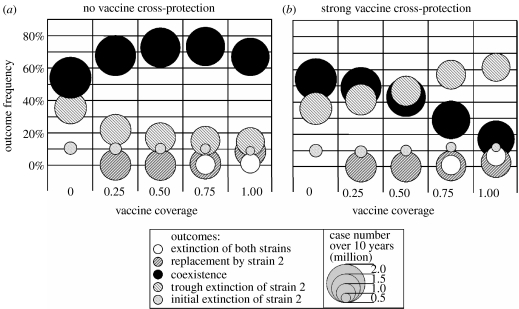
First, we showed that increasing the coverage of a vaccine that confers weak cross-protection would favour the initial spread of a novel strain (figure 2a), and then slightly increase the depth of the trough (figure 4a). Actually, as shown in figure 6a, a vaccine that does not confer protection against a novel strain will enhance its persistence as coverage increases. As expected, the pattern is reversed if the vaccine is more protective than natural immunity (figure 6b); strain 2 still produces an outbreak in about 90% of simulations, but it is more likely to go extinct afterwards as coverage increases. Strain extinction appears to be the main factor affecting disease incidence after the introduction of the second strain. We recorded the total number of cases occurring in the 10 years that follow introduction (represented by the diameter of symbols in figure 6). Over that period, disease incidence is mainly dependent on the occurrence of extinctions. Thus, increasing the coverage of a vaccine that does not confer cross-protection (figure 6a) would have little impact on case numbers in the most probable scenario where both strains coexist. By contrast, increasing the uptake of a cross-protective vaccine (figure 6b) would favour extinctions of strain 2, resulting in lower incidence.
Figure 6.
Summary of 10 series of 1000 stochastic simulations, for two values of vaccine cross-protection ((a) τ=0, (b) τ=0.7) and five values of vaccine coverage (horizontal axis). In each series, we recorded the frequencies (vertical scale) of the five possible outcomes (pattern coded) and the corresponding number of cases (the diameter of bubbles represents the average number for each outcome) in the 10 years that follow the introduction of strain 2. Other parameters are as in figure 5, except natural cross-protection θ=0.5.
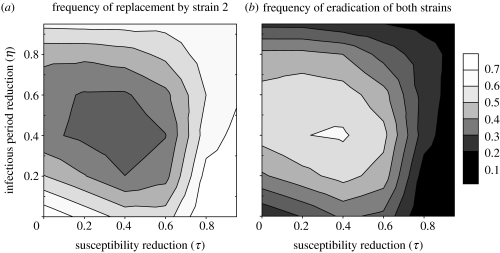
The non-monotonic effects of vaccine cross-protection (τ and η), which we observed with deterministic simulations (figure 4b), are confirmed with stochastic simulations. Indeed, there appears an optimum level (around τ=η=0.4 in figure 7) that maximizes the frequency of trough extinctions, and therefore minimizes average disease incidence after the outbreak (not shown). In other words, novel strains that are most likely to persist (possibly excluding their pre-existing competitors) are either those that are least affected by vaccine immunity (very low τ and η) or those that only marginally escape vaccine immunity (τ and η close to 1). As explained earlier, the latter strategy can be viewed as a prudential use of resources that prevents post-epidemic depletion of susceptible individuals. Conversely, from the pathogen's point of view, stochastic simulations (figure 7) confirm that maintaining infectious period (i.e. low η) is more efficient than maintaining infectiousness (i.e. low τ). In addition, combining intermediate levels of the two forms of evasion appears to be the worst strategy to persist in the population after the initial outbreak.
Figure 7.
(a) Frequencies of replacement of strain 1 by strain 2 and (b) frequencies of eradication of both strains, estimated from 36 series of 1000 stochastic simulations for a range of vaccine cross-protection parameters (τ and η) from 0 to 0.95. Other parameter values are as in figure 5, except natural cross-protection θ=0.6 and ν=0.7. Only three outcomes were obtained in these series: initial extinction of strain 2 (not shown); (a) extinction of strain 1 only; (b) or extinction of both strains.
4. Discussion
As described in §1, most of the recent or prospective immunization campaigns are likely to be challenged by the emergence of antigenically diverse strains. Public health officers need quantitative predictions about the shorter- and longer-term dynamics of those strains as well as the implications for disease epidemiology, in order to derive optimal strategies. However, with a few exceptions (e.g. Stollenwerk et al. 2004), existing models for strain dynamics provide little information on disease dynamics. By combining deterministic and stochastic simulations of a simple model for two strains and a vaccine, we have overcome some of the limitations of classical approaches. The main message from our study is that vaccination not only affects the emergence of antigenic variants, but also their persistence, sometimes in opposite ways.
Here, we focused on a few parameters representing key properties of the vaccine and chose numerical values tailored to pertussis. Although the model would need to be adjusted to obtain predictions on specific diseases and strains, we can derive a few general patterns.
4.1 Strain dynamics
First, we considered the effects of increasing vaccine coverage. In the absence of antigenic variation, this would help fight the disease (see equation (3.1)). However, previous studies (McLean 1995; Lipsitch 1997; White et al. 1998) showed that increasing the coverage of a vaccine with low cross-protection could favour the spread of escape mutants. We confirmed that trend and went on to show that the invasive strain may also be less likely to go extinct after an outbreak as the coverage increases. Conversely, if the vaccine confers higher cross-protection than natural infection, then the frequency of extinction increases with coverage. In summary, a vaccine with narrow antigenic specificity is all the more prone to antigenic evasion, as it is more efficient against the targeted strain.
Second, we varied the strength of cross-protection conferred by the vaccine and found less intuitive patterns. One of the unexpected results was that a high level of vaccine cross-protection (typically, susceptibility to a new strain reduced by 90%) can actually favour the persistence of that new strain. Actually, a vaccine achieving only 50% cross-protection can be more successful in driving the new strain extinct. As we argued earlier, strong cross-protection results in a milder outbreak of the new strain, which prevents a catastrophic depletion of susceptible persons.
Finally, we combined two forms of cross-protection: either reduction in susceptibility or shorter infectious period. Whereas classical epidemiological models predict that the two parameters equally affect the invasion rate of a pathogen (as the basic reproductive ratio R0 is proportional to their product), we found a significant asymmetry, as in our previous model (Restif & Grenfell 2006). In short, if we compare a vaccine that reduces susceptibility with the one that shortens the infectious period, the latter enhances the initial invasion of a new strain, but then makes it more likely to go extinct. Actually, a vaccine that combines mild reductions in both susceptibility and infectious period is most likely to result in strain extinction.
4.2 Implications for disease control
Although we have so far emphasized strain extinction, a factor overlooked by previous studies, health policies should primarily aim at controlling disease incidence. Therefore, it is important to understand the relation between strain dynamics and disease incidence. For example, Stollenwerk et al. (2004) showed that strain diversity could be responsible for irregular meningococcal outbreaks. In this study, we recorded the total number of cases in the 10 years that follow the introduction of the novel strain. Overall, the most favourable outcomes are either an initial extinction of the new strain or later extinction of both strains. However, they amount to very different dynamics: the former results in lower incidence in the short term, but strain 1 remains endemic, whereas the latter ensures long-term elimination of the disease, but only after a big outbreak. If only one strain remains present after the outbreak, the incidence is lower than if both coexist. This appears to be caused by out-of-phase cycles of coexisting strains (as illustrated in figure 5), although this mechanism would require further analyses, as it may depend on the precise nature of interactions between strains (see Gupta et al. 1998; Rohani et al. 1999).
Thus, our method allows the quantification of the probable outcomes of vaccine evasion, both in terms of strain dynamics and expected case numbers. This can help decide on vaccine policies, for example setting up targets for vaccine coverage or choosing between different vaccines with different properties. Other factors, such as population structure or boosters, could be added into the model.
We showed that both the strength and the nature of vaccine cross-protection had important and sometimes counter-intuitive effects. However, estimating these parameters for a given system is feasible only if all strains are already present. The protection against new strains that may arise by mutation is hard to know in advance. Antigenic mapping is a new tool that helps quantify cross-protection in highly diverse pathogens such as human influenza viruses (Smith et al. 2004), but its predictive value is still limited by the knowledge on the genetic and antigenic constraints that may drive pathogen evolution.
Our method provides information on the possible fate of a new mutant, depending on its antigenic distance to the former strain and the vaccine. Interestingly, we found that a very short distance (i.e. strong cross-protection) may promote the spread and persistence of the mutant (see §3.2 for a description of this prudential strategy), although long antigenic distances (i.e. weak cross-protection) are still more favourable.
4.3 Rethinking immune evasion and pathogen evolution
The study of antigenic variation and its implications for disease control often appears to distinguish two mechanisms. On the one hand, some pathogens have evolved huge antigenic diversity in the absence of vaccination, under the pressure of strain competition either within hosts (e.g. HIV) or among them (e.g. human influenza). Here, the main issue is to produce vaccines that are able to protect against a wide range of variants. On the other hand, the very use of vaccines has selected the emergence of novel antigenic mutants (e.g. hepatitis B) or favoured the spread of once rare serotypes (e.g. S. pneumoniae). Such a change raises the question of whether the composition or the use of the vaccine at stake should be updated. This dichotomy is reflected in the theoretical literature on the subject. The former mechanism has been described by models, in which each strain induces specific immune memory that may partly protect against secondary infections by competing strains (e.g. Andreasen et al. 1997; Gupta et al. 1998; Ferguson et al. 1999; Gog & Grenfell 2002). The latter mechanism has been addressed by a group of models (McLean 1995; Lipsitch 1997; White et al. 1998), in which an endemic strain targeted by a vaccine is challenged by a single mutant that (i) can infect vaccinated individuals and (ii) has a lower reproductive ratio R0 than the endemic strain. Thus, the mutant cannot invade unless vaccine coverage and specificity are high enough. Our study aims to draw a bridge between those two frameworks: on the one hand, the two strains considered here differ only in the antigenic space—the mutant does not pay a cost for its ability to evade vaccination; on the other hand, the coverage and the specificity of the vaccine do alter the ability of the mutant to persist and replace the endemic strain.
Together with our previous paper (Restif & Grenfell 2006), this study challenges classical modelling frameworks for the evolution of pathogens. First, the interplay between short-term epidemic dynamics and evolutionary processes cannot be ignored. Second, pathogen evolution is not entirely determined by variations in the basic reproductive ratio R0.
4.4 Concluding remarks
The examples shown in this study were based on a simplistic two-strain model with parameter values inspired by pertussis. Unfortunately, the scarcity of data on pertussis strain dynamics and their relative cross-immunity still limits our ability to test predictions from this model. Recently, Lin et al. (2006) published parallel time-series for pertussis case reports and strain prevalence in Taiwan between 1993 and 2004. During that period, they witnessed two episodes of strain replacement which followed epidemic peaks. Those peaks appear far less dramatic than in most of our simulations (although pertussis report rates are widely regarded as very low), which may indicate strong levels of cross-protection between strains, in which case we expect strain replacement to be a probable outcome. As more accurate data are expected to be published, we now intend to refine our approach to make specific predictions about pertussis or other diseases.
Different model structures or numerical values would affect the depth of the troughs and the chances of extinction. The rule of thumb is that factors that help the strains ‘recover’ from the depletion of the susceptible population tend to make the trough less deep and prevent extinction: higher R0; shorter immune period; and longer infectious period (Restif & Grenfell 2006). Eventually, the probabilities of extinctions will strongly depend on the size of the population (Abu-Raddad & Ferguson 2004). Therefore, this study should be seen as a template that can be tailored to any specific disease. By keeping the model simple, we were able to analyse and explain the results of stochastic simulations, but this may prove harder as the complexity of the model increases.
We are now considering further extensions of the model, in particular a metapopulation framework where sub-populations may represent neighbouring cities (with different sizes) or countries (with different vaccine policies) exchanging infected individuals through migration. We have consciously omitted variations in population sizes and migration terms in this paper, as those factors will be explored in detail in a future study. The present model will then serve as a starting point, representing a relatively small, isolated community.
To conclude, we hope that this study will help promote a sound approach to the design of mathematical models for public health: making a model more realistic does not depend so much on the number of compartments and parameters included, as on a careful choice of underlying assumptions and methods.
Acknowledgments
We thank Pej Rohani and four anonymous referees for their helpful comments on a previous version of the manuscript.
Appendix A. Invasion rate of strain 2
Assuming that strain 1 is at the endemic equilibrium described by equation (3.1), the initial invasion dynamics of strain 2 can approximately be written as
| (A1) |
Let X2(t)=I2(t)+J2(t)+IV2(t), and assume that, over a limited interval t0<t<t1, X2(t) approximately follows an exponential growth written as C ext, where C is a positive constant and x is the exponential invasion rate of strain 2. System (A 1) can then be solved as follows:
| (A2) |
where K2, and KV2 are three constants that depend on initial conditions. If x>0, then the first term in each function rapidly becomes negligible. Since we initially assumed that I2(t)+J2(t)+IV2(t)=C ext, then x must satisfy the following relation:
| (A3) |
It can be seen that f is a continuous, decreasing function over [0, ∞]. If cross-immunity is perfect, i.e. θ=τ=1, then (A 3) admits x=0 as a solution; as expected, strain 2 remains virtually constant (after short transient variations). Otherwise, f(0)>1, so (A 3) admits a unique solution x>0.
Note that this proof relies on the assumption that strain 2 initially follows an exponential growth; if so, then the rate is given by solving equation (A 3). We checked numerically, by integrating the full system of differential equations given by equation (2.1) with the software Mathematica, the fit between the initial dynamics of strain 2 and the predicted exponential growth. For example, across the range of parameter values shown in figure 2b, linear regressions of the function log[I2(t)+J2(t)+IV2(t)] over the interval t=[0, 30 days] gave coefficients of variation R2>0.93 (which represents the fraction of the variation of the data explained by the linear regression), supporting the assumption of an initial exponential growth for strain 2. Over the same range of values, the relative error between x, as given by solving equation (A 3), and the actual growth rate obtained by the linear regression was less than 3%.
References
- Abu-Raddad L.J, Ferguson N.M. The impact of cross-immunity, mutation and stochastic extinction on pathogen diversity. Proc. R. Soc. B. 2004;271:2431–2438. doi: 10.1098/rspb.2004.2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson H, Britton T. Stochastic epidemics in dynamic populations: quasi-stationarity and extinction. J. Math. Biol. 2000;41:559–580. doi: 10.1007/s002850000060. [DOI] [PubMed] [Google Scholar]
- Anderson R.M, May R.M. Oxford University Press; Oxford, UK: 1991. Infectious diseases of humans. [Google Scholar]
- Andreasen V, Lin J, Levin S.A. The dynamics of cocirculating influenza strains conferring partial cross-immunity. J. Math. Biol. 1997;35:825–842. doi: 10.1007/s002850050079. [DOI] [PubMed] [Google Scholar]
- Bartlett M.S. Stochastic processes or the statistics of change. Appl. Stat. 1953;2:44–64. doi: 10.2307/2985327. [DOI] [Google Scholar]
- Brook I, Foote P.A, Hausfeld J.N. Frequency of recovery of pathogens causing acute maxillary sinusitis in adults before and after introduction of vaccination of children with the 7-valent pneumococcal vaccine. J. Med. Microbiol. 2006;55:943–946. doi: 10.1099/jmm.0.46346-0. [DOI] [PubMed] [Google Scholar]
- Broutin H, Rohani P, Guégan J.-F, Grenfell B.T, Simondon F. Loss of immunity to pertussis in a rural community in Senegal. Vaccine. 2004;22:594–596. doi: 10.1016/j.vaccine.2003.07.018. [DOI] [PubMed] [Google Scholar]
- Carman W.F, Zanetti A.R, Karayiannis P, Waters J, Manzillo G, Tanzi E, Zuckerman A.J, Thomas H.C. Vaccine-induced escape mutant of hepatitis B virus. Lancet. 1990;336:325–329. doi: 10.1016/0140-6736(90)91874-A. [DOI] [PubMed] [Google Scholar]
- Cassiday P.K, Sanden G.N, Heuvelman K, Mooi F.R, Bisgard K.M, Popovic T. Polymorphism in Bordetella pertussis pertactin and pertussis toxin virulence factors in the United States, 1935–1999. J. Infect. Dis. 2000;182:1402–1408. doi: 10.1086/315881. [DOI] [PubMed] [Google Scholar]
- Chen W.N, Oon C.J. Hepatitis B virus surface antigen (HBsAg) mutants in Singapore adults and vaccinated children with high anti-hepatitis B virus antibody levels but negative for HBsAg. J. Clin. Microbiol. 2000;38:2793–2794. doi: 10.1128/jcm.38.7.2793-2794.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes J.H.P, Gog J. The onset of oscillatory dynamics of multiple disease strains. J. Math. Biol. 2002;45:471–510. doi: 10.1007/s00285-002-0163-9. [DOI] [PubMed] [Google Scholar]
- de Almeida A.E.C.C, de Filippis I, de Abreu A.O, Ferreira D.G, Gemal A.L, Marzochi K.B.F. Occurrence of Haemophilus influenzae strains in three Brazilian states since the introduction of a conjugate Haemophilus influenzae type b vaccine. Braz. J. Med. Biol. Res. 2005;38:777–781. doi: 10.1590/S0100-879X2005000500016. [DOI] [PubMed] [Google Scholar]
- Elbasha E.H, Galvani A.P. Vaccination against multiple HPV types. Math. Biosci. 2005;197:88–117. doi: 10.1016/j.mbs.2005.05.004. [DOI] [PubMed] [Google Scholar]
- Elomaa A, Advani A, Donnelly D, Antila M, Mertsola J, Hallander H.O, He Q. Strain variation among Bordetella pertussis isolates in Finland, where the whole-cell pertussis vaccine has been used for 50 years. J. Clin. Microbiol. 2005;43:3681–3687. doi: 10.1128/JCM.43.8.3681-3687.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito S, Agliardi T, Giammanco A, Faldella G, Cascio A, Bosis S, Friscia O, Clerici M, Principi N. Long-term pertussis-specific immunity after primary vaccination with a combined diphteria, tetanus, tricomponent accellular pertussis, and hepatitis B vaccine in comparison with that after natural infection. Infect. Immun. 2001;69:4516–4520. doi: 10.1128/IAI.69.7.4516-4520.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson N, Anderson R.M, Gupta S. The effect of antibody-dependent enhancement on the transmission dynamics and persistence of multiple-strains pathogens. Proc. Natl Acad. Sci. USA. 1999;96:790–794. doi: 10.1073/pnas.96.2.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingermann M, et al. Differences of circulating Bordetella pertussis population in Argentina from the strain used in vaccine production. Vaccine. 2006;24:3513–3521. doi: 10.1016/j.vaccine.2006.02.026. [DOI] [PubMed] [Google Scholar]
- Flannery B.P, et al. Changes in invasive pneumococcal diseases among HIV-infected adults living in the era of childhood pneumococcal immunization. Ann. Int. Med. 2006;144:1–9. doi: 10.7326/0003-4819-144-1-200601030-00004. [DOI] [PubMed] [Google Scholar]
- Gibson M.A, Bruck J. Efficient exact stochastic simulation of chemical systems with many species and many channels. J. Phys. Chem. A. 2000;104:1876–1889. doi: 10.1021/jp993732q. [DOI] [Google Scholar]
- Gillespie D.T. Exact stochastic simulation of coupled chemical reactions. J. Phys. Chem. 1977;81:2340–2361. doi: 10.1021/j100540a008. [DOI] [Google Scholar]
- Gog J, Grenfell B.T. Dynamics and selection of many-strain pathogens. Proc. Natl Acad. Sci. USA. 2002;99:17 209–17 214. doi: 10.1073/pnas.252512799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gog J.R, Rimmelzwaan G.F, Osterhaus A.D.M.E, Grenfell B.T. Population dynamics of rapid fixation in cytotoxic T lymphocyte escape mutants of influenza A. Proc. Natl Acad. Sci. USA. 2003;100:11 143–11 147. doi: 10.1073/pnas.1830296100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes M.G.M, Medley G.F, Nokes D.J. On the determinants of population structure in antigenically diverse pathogens. Proc. R. Soc. B. 2002;269:227–233. doi: 10.1098/rspb.2001.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenfell B.T, Anderson R.M. Pertussis in England and Wales: an investigation of transmission dynamics and control by mass vaccination. Proc. R. Soc. B. 1989;236:213–252. doi: 10.1098/rspb.1989.0022. [DOI] [PubMed] [Google Scholar]
- Grenfell B.T, Bjørnstad O.N, Kappey J. Travelling waves and spatial hierarchies in measles epidemics. Nature. 2001;414:716–723. doi: 10.1038/414716a. [DOI] [PubMed] [Google Scholar]
- Grenfell B.T, Pybus O.G, Gog J.R, Wood J.L.N, Daly J.M, Mumford J.A, Holmes E.C. Unifying the epidemiological and evolutionary dynamics of pathogens. Science. 2004;303:327–332. doi: 10.1126/science.1090727. [DOI] [PubMed] [Google Scholar]
- Gupta S, Ferguson N.M, Anderson R.M. Vaccination and the population structure of antigenically diverse pathogens that exchange genetic material. Proc. R. Soc. B. 1997;264:1435–1443. doi: 10.1098/rspb.1997.0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Ferguson N, Anderson R.M. Chaos, persistence and evolution of strain structure in antigenically diverse infectious agents. Science. 1998;280:912–915. doi: 10.1126/science.280.5365.912. [DOI] [PubMed] [Google Scholar]
- Gzyl A, Augustynowicz E, Van Loo I.H.M, Slusarczyk J. Temporal nucleotide changes in pertactin and pertussis toxin genes in Bordetella pertussis strains isolated from clinical cases in Poland. Vaccine. 2002;20:299–303. doi: 10.1016/S0264-410X(01)00356-5. [DOI] [PubMed] [Google Scholar]
- Hsu H.Y, Chang M.-H, Liaw S.H, Ni Y.H, Chen H.-L. Changes of hepatitis B surface antigen variants in carrier children before and after universal vaccination in Taiwan. Hepatology. 1999;30:1312–1317. doi: 10.1002/hep.510300511. [DOI] [PubMed] [Google Scholar]
- Karthigesu V.D, Allison L.M.C, Fergusson M, Howard C.R. A hepatitis B variant found in the sera of immunised children induces a conformational change in the HBsAg “a” mutant. J. Med. Virol. 1999;58:346–352. doi: 10.1002/(SICI)1096-9071(199908)58:4%3C346::AID-JMV5%3E3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Keeling M.J, Grenfell B.T. Disease extinction and community size: modeling the persistence of measles. Science. 1997;275:65–67. doi: 10.1126/science.275.5296.65. [DOI] [PubMed] [Google Scholar]
- Kirupaharan N, Allen L.J.S. Coexistence of multiple pathogen strains in stochastic epidemic models with density-dependent mortality. Bull. Math. Biol. 2004;66:841–864. doi: 10.1016/j.bulm.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Lin Y.-C, Yao S.-M, Yan J.-J, Chen Y.-Y, Hsiao M.-J, Chou C.-Y, Su H.-P, Wu H.-S, Li S.-Y. Molecular epidemiology of Bordetella pertussis in Taiwan, 1993e2004: suggests one possible explanation for the outbreak of pertussis in 1997. Microbes Infect. 2006;8:2082–2087. doi: 10.1016/j.micinf.2006.03.019. [DOI] [PubMed] [Google Scholar]
- Lipsitch M. Vaccination against colonizing bacteria with multiple serotypes. Proc. Natl Acad. Sci. USA. 1997;94:6571–6576. doi: 10.1073/pnas.94.12.6571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrantonio P, Spigaglia P, Van Oirschot H, van der Heide H.G.J, Heuvelman K, Stefanelli P, Mooi F.R. Antigenic variants in Bordetella pertussis strains isolated from vaccinated and unvaccinated children. Microbiology. 1999;145:2069–2075. doi: 10.1099/13500872-145-8-2069. [DOI] [PubMed] [Google Scholar]
- Mbelle N, Huebner R.E, Wasas A.D, Kimura A, Chang I, Klugman P. Immunogenicity and impact on nasopharyngeal carriage of a nonavalent pneumococcal conjugate vaccine. J. Infect. Dis. 1999;180:1171–1176. doi: 10.1086/315009. [DOI] [PubMed] [Google Scholar]
- McLean A.R. Vaccination, evolution and changes in the efficacy of vaccines: a theoretical framework. Proc. R. Soc. B. 1995;261:389–393. doi: 10.1098/rspb.1995.0164. [DOI] [PubMed] [Google Scholar]
- Moghadas S.M, Gumel A.B. A mathematical study of a model for childhood diseases with non-permanent immunity. J. Comput. Appl. Math. 2003;157:347–363. doi: 10.1016/S0377-0427(03)00416-3. [DOI] [Google Scholar]
- Mollison D. Spatial contact models for ecological and epidemic spread. J. R. Stat. Soc. B. 1977;39:283–326. [Google Scholar]
- Mooi F.R, Van Oirschot H, Heuvelman K, Van der Heide H.G.J, Gaasrta W, Willems R.J.L. Polymorphism in the Bordetella pertussis virulence factors P.69/pertactin and pertussis toxin in the Netherlands: temporal trends and evidence for vaccine-driven evolution. Infect. Immun. 1998;66:670–675. doi: 10.1128/iai.66.2.670-675.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nåsell I. Stochastic models of some endemic infections. Math. Biosci. 2002;179:1–19. doi: 10.1016/S0025-5564(02)00098-6. [DOI] [PubMed] [Google Scholar]
- Obaro S.K, Adegbola R.A, Banya W.A.S, Greenwood B.M. Carriage of pneumococci after pneumococcal vaccination. Lancet. 1996;348:271–272. doi: 10.1016/S0140-6736(05)65585-7. [DOI] [PubMed] [Google Scholar]
- Packard E.R, Parton R, Coote J.G, Fry N.K. Sequence variation and conservation in virulence-related genes of Bordetella pertussis isolates from the UK. J. Med. Microbiol. 2004;53:355–365. doi: 10.1099/jmm.0.05515-0. [DOI] [PubMed] [Google Scholar]
- Peppler M.S, Kuny S, Nevesinjac A, Rogers C, de Moissac Y.R, Knowles K, Lorange M, de Serres G, Talbot J. Strain variation among Bordetella pertussis isolates from Québec and Alberta provinces of Canada from 1985 to 1994. J. Clin. Microbiol. 2003;41:3344–3347. doi: 10.1128/JCM.41.7.3344-3347.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdue D.G, Bulkow L.R, Gellin B.G, Davidson M, Petersen K.M, Singleton R.J, Parkinson A.J. Invasive Haemophilus influenzae disease in Alaskan residents aged 10 years and older before and after infant vaccination programs. J. Am. Med. Assoc. 2000;283:3089–3094. doi: 10.1001/jama.283.23.3089. [DOI] [PubMed] [Google Scholar]
- Restif O, Grenfell B.T. Integrating life history and cross-immunity into the evolutionary dynamics of pathogens. Proc. R. Soc. B. 2006;273:409–416. doi: 10.1098/rspb.2005.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohani P, Earn D.J.D, Grenfell B.T. Opposite patterns of synchrony in sympatric diseases metapopulations. Science. 1999;286:968–971. doi: 10.1126/science.286.5441.968. [DOI] [PubMed] [Google Scholar]
- Rohani P, Keeling M.J, Grenfell B.T. The interplay between determinism and stochasticity in childhood diseases. Am. Nat. 2002;159:469–481. doi: 10.1086/339467. [DOI] [PubMed] [Google Scholar]
- Smith D.J, Lapedes A.S, de Jong J.C, Bestebroer T.M, Rimmelzwaan G.F, Osterhaus A.D.M.E, Fouchier R.A.M. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305:371–376. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- Stollenwerk N, Maiden M.C.J, Jansen V.A.A. Diversity in pathogenicity can cause outbreaks of meningococcal disease. Proc. Natl Acad. Sci. USA. 2004;101:10 229–10 234. doi: 10.1073/pnas.0400695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang R.S.W, Mubareka S, Sill M.L, Wylie J, Skinner S, Law D.K.S. Invasive Haemophilus influenzae in Manitoba, Canada, in the postvaccination era. J. Clin. Microbiol. 2006;44:1530–1535. doi: 10.1128/JCM.44.4.1530-1535.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Türkyilmaz A.R, Yurdaydin C, Bozdayi A.M. The first identified hepatitis B virus vaccine escape mutation in Turkey. J. Clin. Virol. 2006;35:201–202. doi: 10.1016/j.jcv.2005.09.003. [DOI] [PubMed] [Google Scholar]
- van Herwaarden O.A. Stochastic epidemics: the probability of extinction of an infectious disease at the end of a major outbreak. J. Math. Biol. 1997;35:793–813. doi: 10.1007/s002850050077. [DOI] [PubMed] [Google Scholar]
- Weber C, Boursaux-Eude C, Coralie G, Caro V, Guiso N. Polymorphism of Bordetella pertussis isolates circulating for the last 10 years in France, where a single effective whole-cell vaccine has been used for more than 30 years. J. Clin. Microbiol. 2001;39:4396–4403. doi: 10.1128/JCM.39.12.4396-4403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White L.J, Cox M.J, Medley G.F. Cross immunity and vaccination against multiple microparasite strains. IMA J. Math. Appl. Med. Biol. 1998;15:211–233. [PubMed] [Google Scholar]
- Wilson J.N, Nokes D.J, Carman W.F. The predicted pattern of emergence of vaccine-resistant hepatitis B: a cause for concern? Vaccine. 1999;17:973–978. doi: 10.1016/S0264-410X(98)00313-2. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Auranen K, Eichner M. The influence of competition and vaccination on the coexistence of two pneumococcal serotypes. Epidemiol. Infect. 2004;132:1073–1081. doi: 10.1017/S0950268804002894. [DOI] [PMC free article] [PubMed] [Google Scholar]