Abstract
Epidermal growth factor receptor (EGFR) gene mutations and increased copy numbers are considered as predictors of response to EGFR tyrosine kinase inhibitors (EGFR-TKI) in non-small-cell lung cancer (NSCLC). Lung cancer diagnosis is often based on cytology alone. However, almost all published data on EGFR gene analyses were obtained from biopsies. This study tested the feasibility of EGFR gene analyses on cytological specimens. Eighty-four cytological specimens from NSCLCs were prospectively analysed for EGFR gene mutation in exons 18–21 and EGFR gene copy numbers were evaluated by fluorescence in situ hybridisation (FISH). A FISH-positive result was defined according to the criteria by Cappuzzo et al established for biopsies of NSCLCs. Fluorescence in situ hybridisation results of cytological specimens were compared to the FISH results on matching biopsies (n=33). Initial diagnosis of NSCLC was solely based on cytology in 37 out of 84 (44.0%) patients. Out of 80 NSCLCs, 6 (7.5%) showed EGFR gene mutations. Out of 67 cancers, 45 (67.2%) were FISH positive on cytological specimens. Comparison of FISH showed a FISH-positive result in 21 out of 33 (63.6%) cytological specimens but in only 8 out of 33 (24.2%) matched biopsies. Epidermal growth factor receptor gene analyses are well applicable to cytological specimens. The high FISH-positive rate of NSCLC on cytological specimens contrasts with the low rate on biopsies when previously suggested criteria are used. New criteria for a positive EGFR FISH status to predict response to therapy with EGFR-TKI need to be defined for cytological specimens.
Keywords: non-small-cell lung cancer, EGFR, mutation, FISH, cytology
Despite strong therapeutic efforts, the overall 5-year survival rate of patients with lung cancer is only 15% without having improved over the last 30 years emphasising the need for new therapies (Jemal et al, 2006).
The epidermal growth factor receptor (EGFR) is known to play a role in the development and progression of cancer, and small molecular EGFR tyrosine kinase inhibitors (EGFR-TKI), including gefitinib (Iressa®; Astra Zeneca, Macclesfield, UK) and erlotinib (Tarceva®; OSI Pharmaceuticals Inc., Melville, NY, USA), are available (Arteaga, 2002; Meert et al, 2002; Schlessinger, 2002; Herbst and Bunn, 2003).
Somatic mutations within the tyrosine kinase domain of the EGFR gene prevail in a subset of non-small-cell lung cancers (NSCLC). These mutations are preferentially found in women, east Asians, never smokers and adenocarcinomas, often with a bronchioloalveolar histology (Fukuoka et al, 2003; Kris et al, 2003; Lynch et al, 2004; Miller et al, 2004; Paez et al, 2004; Perez-Soler et al, 2004; Eberhard et al, 2005; Shepherd et al, 2005; Tsao et al, 2005). Initial retrospective studies showed an average response rate to EGFR-TKI of 75% for NSCLC with EGFR mutations that contrasted with a low response rate of <10% for tumours with wild-type EGFR gene (Riely et al, 2006). These results were subsequently confirmed in prospective studies (Riely et al, 2006). Since response to EGFR-TKI is not fully restricted to patients with EGFR-mutated NSCLC suggests that additional molecular mechanism may be involved (Tsao et al, 2005). In fact, a high EGFR gene copy number detected by fluorescence in situ hybridisation (FISH) was also shown to predict improved survival after EGFR-TKI therapy (Cappuzzo et al, 2005; Hirsch et al, 2005).
A considerable fraction of NSCLCs is diagnosed solely by cytology, and the relative paucity of tumour cells on these specimens is a challenge for molecular analyses (Sequist et al, 2007). Since almost all of the published data on EGFR mutation and gene copy number analyses were made on biopsy material, the goal of this study was to test whether such EGFR gene analyses are feasible on cytological specimens of NSCLCs in a diagnostic setting.
MATERIALS AND METHODS
Cytology and biopsy specimens
A consecutive series of 84 cytological specimens with NSCLC diagnosed during November 2004 to January 2006 was entered into the study. Sixty-five specimens were from primary tumours and 19 from regional lymph node metastases of the mediastinum. The specimens included 35 transbronchial fine needle aspirates, 15 bronchial washings, 13 bronchial brushes, 5 bronchoalveolar lavages and 16 pleural effusions.
The specimens were processed according to routine procedures, using Delaunay's solution as a fixative. They were stained according to Papanicolaou and permanently mounted with coverslips.
In 33 patients, a matched biopsy of the NSCLC was available for comparative analysis. Biopsies were fixed in 4% buffered formalin, and paraffin-embedded biopsies were cut into 4 μm sections and stained with haematoxylin and eosin. In 26 of these 33 paired specimens, both cytology and biopsy were from the primary tumour sites (nine bronchial washings with nine bronchial biopsies; six bronchial brushes with four bronchial biopsies, one pneumonectomy and one pleural biopsy; five transbronchial fine needle aspirates of the lung with three bronchial biopsies and two lobectomies; four pleural effusions with two pleural biopsies, one bronchial biopsy and one lung examined at autopsy; two bronchoalveolar lavages with one bronchial biopsy and one pneumonectomy).
Sequence analysis of the EGFR gene
Cancer cells from Papanicolaou-stained cytological specimens and from haematoxylin–eosin-stained tissue sections were selectively dissected under visual control using laser microdissection in combination with a laser pressure catapulting system according to the manufacturer's guidelines (PALM® MicroBeam, Microlaser Technologies GmbH, Bernried, Germany). Laser energy catapulted cells were collected in the cap of a 0.5 ml plastic tube containing 80 μl of 1 × PCR buffer (Applied Biosystems, Foster City, CA, USA). A 20 μl portion of proteinase K was added and incubated overnight at 56°C. The enzyme was inactivated by heating at 95°C for 10 min. In order to avoid recently described false-positive point mutations by polymerase (Marchetti et al, 2006), we performed two independent multiplex PCRs using 5 μl of this solution, each as a template followed by a second multiplex PCR.
The first multiplex PCR contained all forward and reverse primers for the EGFR exons 18, 19, 20 and 21 (Table 1). For the second multiplex PCR with nested and seminested primers (Table 1), 1 μl from the first multiplex PCR was used as a template.
Table 1. Primers used for the EGFR gene mutation analysis for the first and the second PCR.
|
Primers
|
||
|---|---|---|
| EGFR gene | Forward | Reverse |
| Exon 18 | ||
| First PCR | GCATGGTGAGGGCTGAGGTGA | CCCCACCAGACCATGAGAGGC |
| Second PCR | ACCCTTGTCTCTGTGTTCTTGTCCC | GCCCAGCCCAGAGGCCTGTG |
| Exon 19 | ||
| First PCR | TGCCAGTTAACGTCTTCCTTC | CCACACAGCAAAGCAGAAAC |
| Second PCR | AACGTCTTCCTTCTCTCTCTG | CCACACAGCAAAGCAGAAAC |
| Exon 20 | ||
| First PCR | CCACCATGCGAAGCCACACTGA | TCCTTATCTCCCCTCCCCGTATCTC |
| Second PCR | CCATGCGAAGCCACACTGACGT | CCCCTCCCCGTATCTCCCTTCC |
| Exon 21 | ||
| First PCR | AGCTTCTTCCCATGATGATCTGTCC | GGCAGCCTGGTCCCTGGTGTC |
| Second PCR | TCCCATGATGATCTGTCCCTCACA | CAGGAAAATGCTGGCTGACCTAAAG |
EGFR=epidermal growth factor receptor; PCR=polymerase chain reaction.
For the first and the second PCRs, 50 PCR cycles were performed using the hot start AmpliTaq Gold polymerase (Applied Biosystems) under the following conditions: denaturation: 20 s at 95°C, annealing: 10 s at 59°C, elongation: 40 s at 72°C.
After exon amplification, unused primers were cleaned up with ExoSAP-IT (USB Corporation, Cleveland, OH, USA). After inactivating the enzyme at 80°C for 15 min, we used 0.5 μl as template for the sequencing PCR. Sequencing was performed in forward and reverse direction for every exon.
The sequence amplicons were detected by capillary electrophoresis with laser-induced fluorescence detection (3130 Genetic Analyzer, Applied Biosystems/Hitachi Inc., Foster City, CA, USA). The resulting four visualised sequences (chromatograms) per exon were analysed using SeqScape 2.5 Software (Applied Biosystems). The sequence analysis of the EGFR gene was considered evaluable if at least two chromatograms of every exon and at least one in each separate DNA isolate were readable.
Specimen pretreatment and FISH assay
A hybridisation target area of 18 × 18 mm to 24 × 50 mm depending on cellularity was selected and permanently marked with a diamond pen. The exact locations of the carcinoma cells were saved by a relocation software (Mark&Find Module; Carl Zeiss Vision GmbH, Hallbergmoos, Germany) connected to an automated stage (Type 00-24-473-0000; Carl Zeiss AG, Oberkochen, Germany) on a Zeiss Axioplan 2 epifluorescence microscope (Carl Zeiss GmbH Jena, Jena, Germany). Before uncovering and hybridisation, a representative cell group was photographed with a digital camera (AxioCam Color, Type 412–312).
The commercially available LSI EGFR SpectrumOrange/CEP 7 SpectrumGreen dual-colour probe set (Vysis Inc., Abbott Laboratories, Downers Grove, IL, USA) was used. It includes directly labelled DNA FISH probes for the EGFR gene (7p12, SpectrumOrange) and the centromere of chromosome 7 (at 7p11.1–q11, SpectrumGreen). Fluorescence in situ hybridisation on cytological specimens was performed according to the recommendations of the manufacturer with minor modifications, as described previously (Savic et al, 2006).
For FISH on biopsies, we used haematoxylin–eosin-stained tissue sections using our previously described standard FISH protocol (Stadlmann et al, 2006). No separate procedure for destaining was required.
Enumeration of FISH signals
Enumeration of FISH signals was performed at a magnification of × 630 after automatic relocation of the cancer cells by a relocation software in the DAPI single-bandpass filter set on a fluorescence microscope. Nuclear signal enumeration was only performed on cells with clearly defined nuclear borders and clearly visible signals. A maximum of 100 cancer cells were scored.
NSCLCs were defined as FISH positive according to previously defined criteria by Cappuzzo et al (2005): a FISH-positive result was defined as presence of ‘high polysomy’ (⩾4 EGFR gene copies per nucleus in ⩾40% of the analysed cancer cells) or of amplification (presence of tight EGFR gene clusters and a ratio of EGFR gene to chromosome of ⩾2, or ⩾15 EGFR gene copies per nucleus in ⩾10% of the analysed cancer cells).
Data analysis
Statistical tests included two-tailed Fisher's exact, Pearson's χ2 and Wilcoxon signed-rank test. Statistical significance was defined as P<0.05. Statistical analysis was performed with JMP 5.1 software (SAS Institute, Cary, NC, USA). Receiver operating characteristic (ROC) curve analysis was performed using the statistical software R (v. 2.4.0) (Team, 2004).
RESULTS
Patient and tumour characteristics
Clinicopathological data of the 84 patients are summarised in Table 2. Sixty-three (75.0%) patients had an inoperable UICC (International Union Against Cancer) tumour stage IIIB, IV or a recurrence. There was no significant difference in tumour histology and tumour stage between male and female patients.
Table 2. Patient and tumour characteristics of NSCLCS analysed for EGFR mutation and gene copy number.
| Characteristics | Patients (%) |
|---|---|
| Total | 84 |
| UICC stage | |
| I–IIIA | 20 (23.8) |
| IIIB–IV | 58 (69.0) |
| Recurrence | 5 (6.0) |
| No information | 1 (1.2) |
| Cancer type | |
| NSCLC, NOS | 26 (31.0) |
| AC | 51 (60.7) |
| SCC | 6 (7.1) |
| LCNEC | 1 (1.2) |
AC=adenocarcinoma; EGFR=epidermal growth factor receptor; LCNEC=large cell neuroendocrine carcinoma; NSCLC, NOS=non-small-cell lung carcinoma, not otherwise specified; SCC=squamous cell carcinoma; UICC=International Union Against Cancer.
The diagnosis of lung cancer was solely based on cytology in 37 out of 84 (44.0%), and a simultaneous diagnosis of lung cancer by cytology and biopsy was made in 34 out of 84 (40.5%) patients. The remaining 13 (15.5%) NSCLC were initially diagnosed by cytology and subsequently by biopsy obtained within a median of 20 days (ranging 8–54 days) after cytology.
EGFR gene sequencing and mutation status
Results of EGFR DNA mutation analysis and their associations with clinicopathological data of the 84 patients are summarised in Table 3. DNA sequencing was successful in 78 out of 84 (92.9%) NSCLCs analysed from cytological specimens. We found four EGFR gene mutations (one on a fine needle aspirate and three on bronchial washings). Matched biopsies were available in three of the six NSCLCs that were not evaluable for EGFR mutation. Sequence analysis was repeated on these tissue specimens revealing mutations in two of these, including a point mutation in exon 21 (R832H) and a silent mutation in exon 20 (F795F), respectively. The EGFR gene sequence of the third NSCLC was not evaluable from the biopsy specimen either. Comparison of the 78 cases evaluable for EGFR gene sequencing with the 6 non-evaluable ones revealed no significant differences in the mean microdissected tumour cell areas or the estimated cell counts (data not shown).
Table 3. Association between EGFR mutation and FISH status and clinicopathological characteristics of NSCLCs.
|
EGFR mutationa
|
EGFR FISH status
|
|||
|---|---|---|---|---|
| Characteristics | Present (%) | Absent (%) | Positive (%) | Negative (%) |
| Total | 6 (7.5) | 74 (92.5) | 45 (67.2) | 22 (32.8) |
| Gender | ||||
| Male (n=55) | 2 (3.6) | 53 (96.4) | 32 (68.1) | 15 (31.9) |
| Female (n=25) | 4 (16) | 21 (84) | 13 (65) | 7 (35) |
| P | 0.07 | 1.00 | ||
| UICC stage | ||||
| I–IIIA | 2 (11.1) | 16 (88.9) | 7 (50) | 7 (50) |
| IIIB–IV | 3 (5.4) | 53 (94.6) | 35 (74.5) | 12 (25.5) |
| Recurrence | 0 | 5 | 2 (40) | 3 (60) |
| No information | 1 | 1 | ||
| P (I–IIIA vs IIIB–IV) | 0.59 | 0.11 | ||
| Cancer type | ||||
| NSCLC, NOS | 3 (12.5) | 21 (87.5) | 12 (66.7) | 6 (33.3) |
| AC | 3 (6.1) | 46 (93.9) | 30 (66.7) | 15 (33.3) |
| SCC | 0 | 6 | 3 (75) | 1 (25) |
| LCNEC | 0 | 1 | 0 | 0 |
AC=adenocarcinoma; EGFR=epidermal growth factor receptor; FISH=fluorescence in situ hybridisation; LCNEC=large cell neuroendocrine carcinoma; NSCLC, NOS=non-small-cell lung carcinoma, not otherwise specified; SCC=squamous cell carcinoma; UICC=International Union Against Cancer.
Criteria for a positive FISH result: presence of high polysomy (⩾4 EGFR gene copies per nucleus in ⩾40% of the analysed cancer cells) or of amplification (presence of tight EGFR gene clusters and a ratio of EGFR gene to chromosome of ⩾2, or ⩾15 EGFR gene copies per nucleus in ⩾10% of the analysed cancer cells).
The results include two cases in which the EGFR mutation was detected on the biopsy specimens.
In summary, 6 out of 80 (7.5%) NSCLCs showed EGFR mutations: 4 diagnosed on cytological specimens and 2 on biopsy specimens. These mutations included the well-described deletion E746-A750 in exon 19 and the point mutation L858R in exon 21 as well as the previously described point mutation R832H in exon 21 and the insertion ASV 770–772 in exon 20 (Shigematsu et al, 2005; Giaccone et al, 2006).
Four of the six NSCLCs with EGFR mutation were from female patients.
Table 4 shows a summary of NSCLCs EGFR mutations, including mutation types, EGFR FISH results and patient characteristics.
Table 4. NSCLCs with EGFR gene mutation: summary of EGFR mutation types, FISH results and patient characteristics.
| Patient | EGFR mutation | EGFR FISH result | Sex | Age (years) | UICC stage | Diagnosis | Smoking history |
|---|---|---|---|---|---|---|---|
| 1 | Exon 19 del E746-A750 | Positive amplification | Female | 35 | IV | NSCLC | No |
| 2 | Exon 20 Ins ASV 770–772 | Positive amplification | Female | 68 | IV | AC | Yes |
| 3 | Exon 20 F795F | Positive high polysomy | Male | 71 | —a | AC | —a |
| 4 | Exon 21 L858R | Positive high polysomy | Female | 49 | IV | NSCLC | Yes |
| 5 | Exon 21 R832H | Negative | Male | 66 | IIIA | NSCLC | Yes |
| 6 | Exon 21 R837R | Negative | Female | 81 | IIA | NSCLC | No |
AC=adenocarcinoma; del=deletion; EGFR=epidermal growth factor receptor; FISH=fluorescence in situ hybridisation; ins=insertion; NSCLC=non-small-cell lung carcinoma; UICC=International Union Against Cancer.
No information.
Estimate of the minimal number of cells needed for EGFR gene sequencing
In order to evaluate the minimal number of cells for successful EGFR DNA sequence analysis, we compared the quality of DNA chromatograms from exons 18, 19, 20 and 21 of the EGFR gene obtained from 30, 50 and 100 cancer cells. The cells were dissected from five cytological specimens from NSCLC with known EGFR mutation status. Four of these NSCLCs had no EGFR mutation and one had a point mutation in exon 21 (L585R). The best result was obtained with 100 analysed cells with 19 out of 20 evaluable chromatograms, followed by 50 cells with 18 out of 20 evaluable chromatograms. With 30 analysed cancer cells, only 12 chromatograms were evaluable. The differences in the evaluability between the different cell counts were not statistically significant. The point mutation in exon 21 (L858R) was identified with 30, 50 and 100 cells.
EGFR gene copy number
In 75 out of 84 (89.3%) specimens, a sufficient number of cancer cells remained for FISH analysis after tumour cell dissection for EGFR gene mutation analysis. Fluorescence in situ hybridisation analysis was successful in 67 out of 75 (89.3%) specimens. The mean number of scored cells was 70±32.7 (ranging 12–100; 95% confidence interval (CI): 62.0–78.0). Fluorescence in situ hybridisation was positive according to the criteria defined by Cappuzzo et al (2005) in 45 out of 67 (67.1%) of the cytological specimens, including 35 out of 67 (52.2%) NSCLCs with ‘high polysomy’ and 10 out of 67 (14.9%) with amplification.
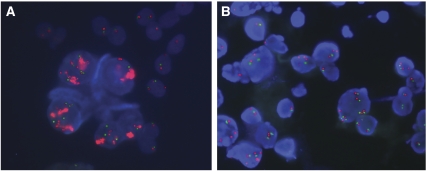
Representative images of FISH-positive results are illustrated in Figure 1.
Figure 1.
Representative images of FISH-positive results on cytological specimens (EGFR gene: red signals; chromosome 7: green signals). (A) Amplification of the EGFR gene with tight EGFR gene clusters; (B) high polysomy of the EGFR gene in the cancer cells. FISH=fluorescence in situ hybridisation; EGFR=epidermal growth factor receptor (see online version for colour figure).
A matched biopsy specimen was available in 33 patients for comparative FISH analysis. The mean number of scored cells was significantly higher in the biopsy specimens compared to the cytology specimens (mean 88.6±17.6: ranging 43–100; 95% CI: 82.4–94.9 vs mean 66.1±31.6: ranging 12–100; 95% CI: 54.9–77.3; P<0.01). Fluorescence in situ hybridisation was positive in 21 (63.6%) of these 33 cytological specimens, including 19 (57.6%) cases with ‘high polysomy’ and 2 (6%) cases with amplification. Fluorescence in situ hybridisation on biopsy specimens was positive in only 8 out of 33 (24.2%) NSCLCs, including 5 (15.1%) with ‘high polysomy’ and 3 (9.1%) with amplification (Table 5). The substantial difference in the FISH results between the matched cytologies and biopsies was almost unchanged when the analysis was restricted to the primary tumours (n=26) after exclusion of the regional lymph node metastases (P=0.01). Thus, the difference cannot be explained by a change of FISH status during progression to metastasis. The number of scored cells remained significantly higher in the biopsy specimens compared to the cytology specimens.
Table 5. Association between EGFR FISH results in the cytological specimens and matched biopsy specimens from all NSCLCs (n=33) and from the subgroup of primary NSCLCs without specimens from regional lymph node metastases (n=26).
| Cytological specimens (%) | Biopsy specimens (%) | P-value | |
|---|---|---|---|
| Total | 33 | 33 | |
| FISH positive | 21 (63.6) | 8 (24.2) | |
| High polysomy | 19 (57.6) | 5 (15.1) | |
| Amplification | 2 (6) | 3 (9.1) | |
| FISH negative | 12 (36.4) | 25 (75.8) | |
| <0.01a | |||
| Primary tumour b | 26 | 26 | |
| FISH positive | 16 (61.5) | 7 (26.9) | |
| High polysomy | 14 (53.8) | 4 (15.4) | |
| Amplification | 2 (7.7) | 3 (11.5) | |
| FISH negative | 10 (38.5) | 19 (73.1) | |
| 0.01a |
EGFR=epidermal growth factor recepetor gene; FISH=fluorescence in situ hybridisation; NSCLC=non-small-cell lung carcinoma.
P-value for difference in FISH positivity between cytology and biopsy.
Both cytology and biopsy from the primary tumour site.
Two amplified NSCLCs showed amplification both in the cytology and in biopsy specimen. In another NSCLC EGFR gene amplification was found in the biopsy but not in the cytological specimen. A review of the slides revealed that the tumour cells in the biopsy were much less differentiated than the cells in the cytological specimen. This suggests that the discrepancy between cytology and histology in this patient could be due to tumour heterogeneity.
In order to synchronise the FISH status of cytological and histological specimens, we tuned the threshold of a positive FISH result for cytological specimens using ROC curve based on the 33 cases with matched specimens (Linden, 2006). Changing the number of gene copies and the minimal percentage of cells in cytological specimens would identify the ideal threshold with maximal sensitivity and specificity. Applying the set of parameters used for definition of ‘high polysomy’ on biopsies (⩾4 copies in ⩾40% of cells) to cytology resulted in a sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV) and κ of 1.0, 0.45, 0.33, 1.0 and 0.26, respectively. The optimised parameters obtained by the ROC curve for ‘high polysomy’ on cytological specimens were ⩾5 copies in ⩾70% of cells. The sensitivity, specificity, PPV, NPV and κ were 0.67, 1.0, 1.0, 0.92 and 0.76 respectively.
Comparison between mutation and FISH status showed that four of the six EGFR gene-mutated NSCLCs had a FISH-positive result, including two with ‘high polysomy’ and two with amplification. Epidermal growth factor receptor gene analysis was mostly performed on cells from Papanicolaou-stained specimens but also worked well in cases in which only immunostained slides were available. The EGFR gene sequence was evaluable in 5 out of 6 specimens and FISH in 7 out of 10 specimens after immunocytochemistry for TTF-1, or BerEp4, or cytokeratine 7, or cytokeratine 20.
DISCUSSION
In this study, we demonstrate that a comprehensive EGFR gene analysis for prediction of response to EGFR-TKI in NSCLC is well feasible in routinely processed cytological specimens. In most patients, small biopsies or cytological specimens must suffice for both morphological diagnosis and predictive marker analyses. Importantly, as in our series, NSCLC is often diagnosed by cytology alone (Rivera et al, 2003). In contrast, most previous studies on EGFR gene mutations or copy number analyses in lung cancer were based on resection specimens or biopsy material. The clinical effect of EGFR-TKI in a proportion of patients with NSCLC is associated with several molecular alterations, including EGFR mutations and increased copy number of the EGFR gene, although the relative importance of the alterations remains controversial (Cappuzzo et al, 2006; Dziadziuszko et al, 2006; Riely et al, 2006).
We found EGFR mutations in 7.5% of the patients. This is in accordance with previous studies in Caucasian patients where the prevalence ranged from 4.5 to 14.1% (Kosaka et al, 2004; Marchetti et al, 2005; Sasaki et al, 2005; Tokumo et al, 2005; Yang et al, 2005).
For mutation analysis, enrichment of morphologically identified cancer cells is crucial in order to avoid dilution of tumour DNA with non-mutated normal DNA from benign cells that often outnumber the malignant cells. This can readily be achieved by laser microdissection. We were able to determine the DNA sequence from as few as 30 tumour cells.
Cappuzzo et al (2005) were the first to propose a strong predictive role of EGFR gene copy number for response to therapy with EGFR-TKI in NSCLC. They found a positive EGFR FISH status defined as ‘high polysomy’ or ‘amplification’ in 32.4% of biopsies from 102 patients with NSCLC being an independent predictor of response to erlotinib. The importance of EGFR gene copy number for therapy response to EGFR-TKI has subsequently been confirmed by several studies (Bell et al, 2005; Eberhard et al, 2005; Tsao et al, 2005; Hirsch et al, 2006). However, all of these previous studies examining EGFR FISH were based solely on histological tissue sections and did not consider cytological specimens. This may have been due to difficulties in differentiating reactive cells from admixed cancer cells after hybridisation. In fact, we regard automated relocation of carcinoma cells identified in Papanicolaou-stained specimens as a prerequisite of FISH analysis in cases in which cancer cells are admixed with a high proportion of benign cells. Disregarding cytological specimens in a previous randomised study on erlotinib in 731 NSCLCs may be one of the reasons why the percentage of specimens used for mutation and FISH analyses was as low as 27 and 30%, respectively (Shepherd et al, 2005; Tsao et al, 2005).
Applying the criteria of Cappuzzo et al in our study revealed a high FISH-positive rate of NSCLC in cytological specimens (63.6%) that contrasted with a low rate in matched biopsy specimens (24.2%). The latter was in line with the prevalence of FISH positivity found in previous studies (Table 6). The discrepancy between cytology and histology is explained by the fact that a proportion of the cell nuclei on biopsy tissue sections are truncated, leading to a lower number of gene signals per nucleus. In contrast, the cell nuclei on the cytological specimens remain intact and contain the true number of gene copies. The discrepancy remained almost unchanged when analysing only primary tumours, dispelling gene copy differences due to tumour heterogeneity between primary tumours and locoregional metastases. Thus, thresholds of FISH positivity defined on histological sections cannot directly be translated to cytological specimens. This is especially true for low-copy-number gains such as the category of ‘high polysomy’, where at least four signals per nucleus are required in at least 40% of the cells. A proportion of tumour cells with three signals or two signals in histological sections would in fact contain four signals if the nuclei were not truncated. Thus, some of these cases would cross the threshold for FISH positivity if the nuclei were intact. In contrast, nuclear truncation does not affect the ability to detect high-level amplifications, as previously shown in cytohistological comparisons of HER-2 FISH analysis in breast cancer (Bozzetti et al, 2002; Gu et al, 2005). This concurs with our results where the difference between histology and cytology was greatest in the category of ‘high polysomy (57.6 vs 15.2%, P<0.01), whereas the prevalence of amplification was comparably low (6 vs 9.1%). An optimised threshold of ⩾5 gene copies in ⩾70% of cells as calculated from ROC curve analysis could substantially improve the specificity and PPV of FISH in cytology when the FISH result of histology was used as a gold standard. However, this was at the cost of reduced sensitivity to a low 67%. There have been previous attempts to compensate for the discrepancies between whole-cell preparations and tissue sections for more accurate enumeration of FISH signals (Berezuk et al, 2001). However, the proposed correction factors based on the cell diameter are limited by the variability of nuclear size and shape in cancer cells, which makes this approach insufficient in a diagnostic setting.
Table 6. Comparison of published EGFR FISH results using the criteria proposed by Cappuzzo et al (2005) with present data.
| Study | Number of evaluated NSCLC | Number of scored cells | FISH positive (%) | Balanced (‘high’) polysomy (%) | Amplification (%) |
|---|---|---|---|---|---|
| Cappuzzo et al (2005) a | 102 | —b | 33 | 20 | 13 |
| Tsao et al (2005) a | 125 | 33–100 | 45 | 34 | 11 |
| Hirsch et al (2005) a | 81 | ⩾100 | 32 | —b | —b |
| Hirsch et al (2006) a | 370 | —b | 31 | 17 | 14 |
| Jeon et al (2006) a | 262 | —b | 30.2 | 19.1 | 11.1 |
| Ichihara et al (2007) a | 75 | ⩾100 | 41.3 | 25.3 | 16 |
| Present study | |||||
| Histology | 33 | 43–100 (mean: 88.6±17.6) | 24.2 | 15.1 | 9.1 |
| Cytology | 67 | 12–100 (mean: 66.1±31.6) | 67.1 | 52.2 | 14.9 |
EGFR=epidermal growth factor receptor gene; FISH=fluorescence in situ hybridisation; NSCLC=non-small-cell lung carcinoma.
Criteria for a positive FISH result: presence of high polysomy (⩾4 EGFR gene copies per nucleus in ⩾40% of the analysed cancer cells) or of amplification (presence of tight EGFR gene clusters and a ratio of EGFR gene to chromosome of ⩾2, or ⩾15 EGFR gene copies per nucleus in ⩾10% of the analysed cancer cells).
All cited FISH analyses were performed on histological specimens.
No information.
Cytological specimens and matched biopsies are not always from the identical tumour area. Therefore, it is possible that the difference in EGFR FISH status between cytology and biopsy in this study might be partly attributable to heterogeneity. However, it is very unlikely that tumour heterogeneity had a unidirectional effect towards a 3.8-fold higher rate of ‘high polysomy’ in cytology than in biopsy (57.6 vs 15.1%). It is similarly unlikely that the lower mean number of tumour cells available for EGFR FISH analysis in cytological specimens selected for ‘high polysomy’. A study on a larger series of matched histological and cytological specimens is under way to investigate the impact of tumour heterogeneity and cell count on the EGFR FISH status, and to define better criteria of a positive EGFR FISH result in cytological specimens.
It is becoming increasingly important to analyse prognostic and predictive markers in tumour biopsies as exemplified by EGFR gene analysis in NSCLC. This is also true for cytological specimens that are often the only source of cells from primary tumours or distant metastatic sites. When dealing with limited cell material in a diagnostic setting, the morphological diagnosis still has priority over molecular analyses. Here, we show that comprehensive molecular data can be obtained even from few cancer cells on diagnostic cytological specimens, which is facilitated by laser microdissection and automated relocation. Our data challenge the recently expressed opinion that the ‘widespread use of mutational analysis is currently hindered by the routine use of very small fragments of tissue to establish the diagnosis of NSCLC’ (Riely et al, 2006). Importantly, standard staining such as Papanicolaou does not interfere with FISH and DNA analysis by PCR, and these methods can even be applied to previously immunostained specimens. This is important since immunocytochemical assessment of proteins such as EGFR or p-Akt may also be of value in predicting response to EGFR-TKI (Cappuzzo et al, 2004; Tsao et al, 2005). Notably, these techniques and tools are not restricted to EGFR analysis and also set the stage for the analysis of future therapeutic targets.
Footnotes
DISCLOSURE
LB received speakers honorary from Abbott. The other authors do not have a pertinent financial relationship with a commercial entity that has an interest in the subject of this paper. This study was in accordance with the guidelines of the ethical committee of the University of Basel.
References
- Arteaga CL (2002) Overview of epidermal growth factor receptor biology and its role as a therapeutic target in human neoplasia. Semin Oncol 29: 3–9 [DOI] [PubMed] [Google Scholar]
- Bell DW, Lynch TJ, Haserlat SM, Harris PL, Okimoto RA, Brannigan BW, Sgroi DC, Muir B, Riemenschneider MJ, Iacona RB, Krebs AD, Johnson DH, Giaccone G, Herbst RS, Manegold C, Fukuoka M, Kris MG, Baselga J, Ochs JS, Haber DA (2005) Epidermal growth factor receptor mutations and gene amplification in non-small-cell lung cancer: molecular analysis of the IDEAL/INTACT gefitinib trials. J Clin Oncol 23: 8081–8092 [DOI] [PubMed] [Google Scholar]
- Berezuk M, West J, Varella-Garcia M, Franklin WA (2001) Adjusting interphase FISH results in epithelial tissue sections to whole cell complement. Anal Quant Cytol Histol 23: 93–100 [PubMed] [Google Scholar]
- Bozzetti C, Nizzoli R, Guazzi A, Flora M, Bassano C, Crafa P, Naldi N, Cascinu S (2002) HER-2/neu amplification detected by fluorescence in situ hybridization in fine needle aspirates from primary breast cancer. Ann Oncol 13: 1398–1403 [DOI] [PubMed] [Google Scholar]
- Cappuzzo F, Finocchiaro G, Metro G, Bartolini S, Magrini E, Cancellieri A, Trisolini R, Castaldini L, Tallini G, Crino L (2006) Clinical experience with gefitinib: an update. Crit Rev Oncol Hematol 58: 31–45 [DOI] [PubMed] [Google Scholar]
- Cappuzzo F, Hirsch FR, Rossi E, Bartolini S, Ceresoli GL, Bemis L, Haney J, Witta S, Danenberg K, Domenichini I, Ludovini V, Magrini E, Gregorc V, Doglioni C, Sidoni A, Tonato M, Franklin WA, Crino L, Bunn Jr PA, Varella-Garcia M (2005) Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non-small-cell lung cancer. J Natl Cancer Inst 97: 643–655 [DOI] [PubMed] [Google Scholar]
- Cappuzzo F, Magrini E, Ceresoli GL, Bartolini S, Rossi E, Ludovini V, Gregorc V, Ligorio C, Cancellieri A, Damiani S, Spreafico A, Paties CT, Lombardo L, Calandri C, Bellezza G, Tonato M, Crino L (2004) Akt phosphorylation and gefitinib efficacy in patients with advanced non-small-cell lung cancer. J Natl Cancer Inst 96: 1133–1141 [DOI] [PubMed] [Google Scholar]
- Dziadziuszko R, Hirsch FR, Varella-Garcia M, Bunn Jr PA (2006) Selecting lung cancer patients for treatment with epidermal growth factor receptor tyrosine kinase inhibitors by immunohistochemistry and fluorescence in situ hybridization – why, when, and how? Clin Cancer Res 12: 4409s–4415s [DOI] [PubMed] [Google Scholar]
- Eberhard DA, Johnson BE, Amler LC, Goddard AD, Heldens SL, Herbst RS, Ince WL, Janne PA, Januario T, Johnson DH, Klein P, Miller VA, Ostland MA, Ramies DA, Sebisanovic D, Stinson JA, Zhang YR, Seshagiri S, Hillan KJ (2005) Mutations in the epidermal growth factor receptor and in KRAS are predictive and prognostic indicators in patients with non-small-cell lung cancer treated with chemotherapy alone and in combination with erlotinib. J Clin Oncol 23: 5900–5909 [DOI] [PubMed] [Google Scholar]
- Fukuoka M, Yano S, Giaccone G, Tamura T, Nakagawa K, Douillard JY, Nishiwaki Y, Vansteenkiste J, Kudoh S, Rischin D, Eek R, Horai T, Noda K, Takata I, Smit E, Averbuch S, Macleod A, Feyereislova A, Dong RP, Baselga J (2003) Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced non-small-cell lung cancer (The IDEAL 1 Trial) [corrected]. J Clin Oncol 21: 2237–2246 [DOI] [PubMed] [Google Scholar]
- Giaccone G, Gallegos Ruiz M, Le Chevalier T, Thatcher N, Smit E, Rodriguez JA, Janne P, Oulid-Aissa D, Soria JC (2006) Erlotinib for frontline treatment of advanced non-small cell lung cancer: a phase II study. Clin Cancer Res 12: 6049–6055 [DOI] [PubMed] [Google Scholar]
- Gu M, Ghafari S, Zhao M (2005) Fluorescence in situ hybridization for HER-2/neu amplification of breast carcinoma in archival fine needle aspiration biopsy specimens. Acta Cytol 49: 471–476 [DOI] [PubMed] [Google Scholar]
- Herbst RS, Bunn Jr PA (2003) Targeting the epidermal growth factor receptor in non-small cell lung cancer. Clin Cancer Res 9: 5813–5824 [PubMed] [Google Scholar]
- Hirsch FR, Varella-Garcia M, Bunn Jr PA, Franklin WA, Dziadziuszko R, Thatcher N, Chang A, Parikh P, Pereira JR, Ciuleanu T, von Pawel J, Watkins C, Flannery A, Ellison G, Donald E, Knight L, Parums D, Botwood N, Holloway B (2006) Molecular predictors of outcome with gefitinib in a phase III placebo–controlled study in advanced non-small-cell lung cancer. J Clin Oncol 24: 5034–5042 [DOI] [PubMed] [Google Scholar]
- Hirsch FR, Varella-Garcia M, McCoy J, West H, Xavier AC, Gumerlock P, Bunn Jr PA, Franklin WA, Crowley J, Gandara DR (2005) Increased epidermal growth factor receptor gene copy number detected by fluorescence in situ hybridization associates with increased sensitivity to gefitinib in patients with bronchioloalveolar carcinoma subtypes: a Southwest Oncology Group Study. J Clin Oncol 23: 6838–6845 [DOI] [PubMed] [Google Scholar]
- Ichihara S, Toyooka S, Fujiwara Y, Hotta K, Shigematsu H, Tokumo M, Soh J, Asano H, Ichimura K, Aoe K, Aoe M, Kiura K, Shimizu K, Date H, Shimizu N (2007) The impact of epidermal growth factor receptor gene status on gefitinib-treated Japanese patients with non-small cell lung cancer. Int J Cancer 120: 1239–1247 [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ (2006) Cancer statistics, 2006. CA Cancer J Clin 56: 106–130 [DOI] [PubMed] [Google Scholar]
- Jeon YK, Sung S, Chung J, Park W, Seo J, Kim CW, Chung DH (2006) Clinicopathologic features and prognostic implications of epidermal growth factor receptor (EGFR) gene copy number and protein expression in non-small cell lung cancer. Lung Cancer 54: 387–398 [DOI] [PubMed] [Google Scholar]
- Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T (2004) Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res 64: 8919–8923 [DOI] [PubMed] [Google Scholar]
- Kris MG, Natale RB, Herbst RS, Lynch Jr TJ, Prager D, Belani CP, Schiller JH, Kelly K, Spiridonidis H, Sandler A, Albain KS, Cella D, Wolf MK, Averbuch SD, Ochs JJ, Kay AC (2003) Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non-small cell lung cancer: a randomized trial. JAMA 290: 2149–2158 [DOI] [PubMed] [Google Scholar]
- Linden A (2006) Measuring diagnostic and predictive accuracy in disease management: an introduction to receiver operating characteristic (ROC) analysis. J Eval Clin Pract 12: 132–139 [DOI] [PubMed] [Google Scholar]
- Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA (2004) Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 350: 2129–2139 [DOI] [PubMed] [Google Scholar]
- Marchetti A, Felicioni L, Buttitta F (2006) Assessing EGFR mutations. N Engl J Med 354: 526–528; author reply 526–528 [DOI] [PubMed] [Google Scholar]
- Marchetti A, Martella C, Felicioni L, Barassi F, Salvatore S, Chella A, Camplese PP, Iarussi T, Mucilli F, Mezzetti A, Cuccurullo F, Sacco R, Buttitta F (2005) EGFR mutations in non-small-cell lung cancer: analysis of a large series of cases and development of a rapid and sensitive method for diagnostic screening with potential implications on pharmacologic treatment. J Clin Oncol 23: 857–865 [DOI] [PubMed] [Google Scholar]
- Meert AP, Martin B, Delmotte P, Berghmans T, Lafitte JJ, Mascaux C, Paesmans M, Steels E, Verdebout JM, Sculier JP (2002) The role of EGF-R expression on patient survival in lung cancer: a systematic review with meta-analysis. Eur Respir J 20: 975–981 [DOI] [PubMed] [Google Scholar]
- Miller VA, Kris MG, Shah N, Patel J, Azzoli C, Gomez J, Krug LM, Pao W, Rizvi N, Pizzo B, Tyson L, Venkatraman E, Ben-Porat L, Memoli N, Zakowski M, Rusch V, Heelan RT (2004) Bronchioloalveolar pathologic subtype and smoking history predict sensitivity to gefitinib in advanced non-small-cell lung cancer. J Clin Oncol 22: 1103–1109 [DOI] [PubMed] [Google Scholar]
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M (2004) EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 304: 1497–1500 [DOI] [PubMed] [Google Scholar]
- Perez-Soler R, Chachoua A, Hammond LA, Rowinsky EK, Huberman M, Karp D, Rigas J, Clark GM, Santabarbara P, Bonomi P (2004) Determinants of tumor response and survival with erlotinib in patients with non-small-cell lung cancer. J Clin Oncol 22: 3238–3247 [DOI] [PubMed] [Google Scholar]
- Riely GJ, Politi KA, Miller VA, Pao W (2006) Update on epidermal growth factor receptor mutations in non-small cell lung cancer. Clin Cancer Res 12: 7232–7241 [DOI] [PubMed] [Google Scholar]
- Rivera MP, Detterbeck F, Mehta AC (2003) Diagnosis of lung cancer: the guidelines. Chest 123: 129S–136S [DOI] [PubMed] [Google Scholar]
- Sasaki H, Endo K, Konishi A, Takada M, Kawahara M, Iuchi K, Matsumura A, Okumura M, Tanaka H, Kawaguchi T, Shimizu T, Takeuchi H, Yano M, Fukai I, Fujii Y (2005) EGFR mutation status in Japanese lung cancer patients: genotyping analysis using LightCycler. Clin Cancer Res 11: 2924–2929 [DOI] [PubMed] [Google Scholar]
- Savic S, Glatz K, Schoenegg R, Spieler P, Feichter G, Tamm M, Bubendorf L (2006) Multitarget fluorescence in situ hybridization elucidates equivocal lung cytology. Chest 129: 1629–1635 [DOI] [PubMed] [Google Scholar]
- Schlessinger J (2002) Ligand-induced, receptor-mediated dimerization and activation of EGF receptor. Cell 110: 669–672 [DOI] [PubMed] [Google Scholar]
- Sequist LV, Bell DW, Lynch TJ, Haber DA (2007) Molecular predictors of response to epidermal growth factor receptor antagonists in non-small-cell lung cancer. J Clin Oncol 25: 587–595 [DOI] [PubMed] [Google Scholar]
- Shepherd FA, Rodrigues Pereira J, Ciuleanu T, Tan EH, Hirsh V, Thongprasert S, Campos D, Maoleekoonpiroj S, Smylie M, Martins R, van Kooten M, Dediu M, Findlay B, Tu D, Johnston D, Bezjak A, Clark G, Santabarbara P, Seymour L (2005) Erlotinib in previously treated non-small-cell lung cancer. N Engl J Med 353: 123–132 [DOI] [PubMed] [Google Scholar]
- Shigematsu H, Lin L, Takahashi T, Nomura M, Suzuki M, Wistuba II, Fong KM, Lee H, Toyooka S, Shimizu N, Fujisawa T, Feng Z, Roth JA, Herz J, Minna JD, Gazdar AF (2005) Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J Natl Cancer Inst 97: 339–346 [DOI] [PubMed] [Google Scholar]
- Stadlmann S, Gueth U, Reiser U, Diener PA, Zeimet AG, Wight E, Mirlacher M, Sauter G, Mihatsch MJ, Singer G (2006) Epithelial growth factor receptor status in primary and recurrent ovarian cancer. Mod Pathol 19: 607–610 [DOI] [PubMed] [Google Scholar]
- Team RDC (2004) R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing [Google Scholar]
- Tokumo M, Toyooka S, Kiura K, Shigematsu H, Tomii K, Aoe M, Ichimura K, Tsuda T, Yano M, Tsukuda K, Tabata M, Ueoka H, Tanimoto M, Date H, Gazdar AF, Shimizu N (2005) The relationship between epidermal growth factor receptor mutations and clinicopathologic features in non-small cell lung cancers. Clin Cancer Res 11: 1167–1173 [PubMed] [Google Scholar]
- Tsao MS, Sakurada A, Cutz JC, Zhu CQ, Kamel-Reid S, Squire J, Lorimer I, Zhang T, Liu N, Daneshmand M, Marrano P, da Cunha Santos G, Lagarde A, Richardson F, Seymour L, Whitehead M, Ding K, Pater J, Shepherd FA (2005) Erlotinib in lung cancer – molecular and clinical predictors of outcome. N Engl J Med 353: 133–144 [DOI] [PubMed] [Google Scholar]
- Yang SH, Mechanic LE, Yang P, Landi MT, Bowman ED, Wampfler J, Meerzaman D, Hong KM, Mann F, Dracheva T, Fukuoka J, Travis W, Caporaso NE, Harris CC, Jen J (2005) Mutations in the tyrosine kinase domain of the epidermal growth factor receptor in non-small cell lung cancer. Clin Cancer Res 11: 2106–2110 [DOI] [PubMed] [Google Scholar]