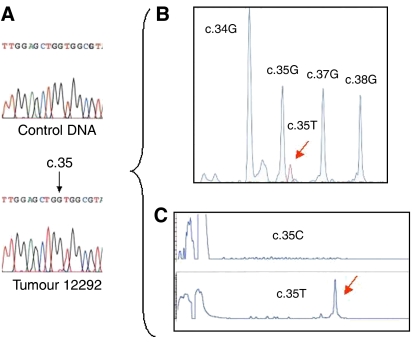
Figure 1.
Detection by SNaPShot and PCR-LCR assays of KRAS mutations not detected by direct sequencing. Direct sequencing of KRAS exon 2 from control DNA and tumour 12292. The black arrow indicates the nucleotide c.35 (A). SNaPShot detection of the c.35G>T (p.G12V) mutation in tumour 12292. Each peak corresponds to a specific extended primer. The red arrows indicates the peak specific of the c.35G>T mutation (B). PCR-LCR analysis of tumour 12292, using a dye-labelled primer specific for the mutant c.35G>C (p.G12A) or c.35G>T (p.G12V) KRAS allele. The arrow indicates the peak specific of the c.35G>T mutation (C). Note that the c.35G>T mutation detected by both the SNaPShot and PCR-LCR assays cannot be clearly detected by sequencing analysis alone.