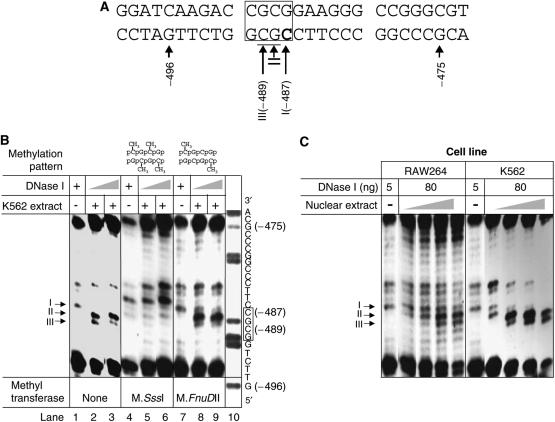
Figure 2.
Footprint 1 site (Fp1). (A) Sequence of the footprint 1 site. The ciphers I, II and III correspond to the footprinting positions from the DNase I protection assay. The DNase I-protected position is shown in bold and DNase I-hypersensitive positions are underlined. The recognition sequence of M.FnuDII is boxed. (B) DNase I protection assays. Methylation of the promoter fragment modifies binding of nuclear extract proteins to footprint 1 site. Compared to the promoter fragments partially digested by DNase I alone (lane 1), addition of nuclear extracts (90 μg) from K562 cells changed the digestion pattern. While one signal (I) is reduced, the intensity of two other bands is increased (II and III). M.SssI methylates every cytosine residue in a CpG dinucleotide, and this methylation abolishes binding of trans-acting factors (lanes 4–6 are essentially identical). M.FnuDII only methylates the lateral cytosin residues in a CpG tetranucleotide and apparently this hemimethylation does not inhibit protective binding of trans-acting factors (lanes 7–9 are essentially identical to lanes 1–3). The A+G ladder (lane 10) shows the reverse complementary sequence. Numbers refer to positions relative to A in the START-ATG (=+1); the M.FnuDII recognition sequence is boxed. (C) DNase I protection assays: The binding of the trans-acting factors is not cell specific. DNase I protection assay with increasing amounts (20, 50, 90 and 120 μg) of nuclear extracts. Band I loses intensity as the amount of nuclear extracts is increased, whereas bands II and III gain intensity. Thus, band I is protected by protein binding and bands II and III are hypersensitive to DNase digestion by protein binding, regardless of the source of the nuclear extract.