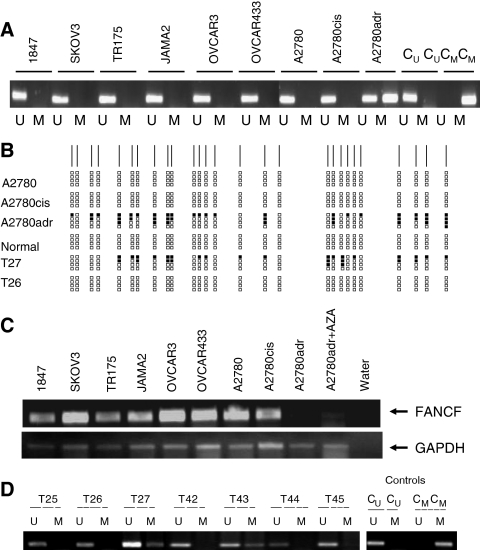
Figure 1.
Methylation analysis of the FANCF CpG island in epithelial ovarian cancer. (A) Methylation-sensitive PCR analysis of the FANCF CpG island in ovarian cancer cell lines. Methylation-sensitive PCR was performed in the indicated cell lines as described in the Materials and Methods. For each cell line, unmethylated (U) and methylated (M) reactions are shown. Control unmethylated (CU) and methylated (CM) reactions are also shown. (B) Bisulphite sequencing of part of the FANCF CpG island. The CpG sites are shown as vertical lines on the top horizontal line. Methylated CpG sites are shown as black blocks. Five levels of methylation are indicated: 0 – no black blocks; 1–25% – one black block; 25–50% – two black blocks; 50–75% – three black blocks; and 75–100% – four black blocks. (C) Reverse transcription-polymerase chain reaction analysis of FANCF expression in epithelial ovarian cancer cell lines. Reverse transcription-polymerase chain reaction was performed in the indicated cell lines as described in the Materials and Methods. In the case of A2780adr, reactivation of expression by AZA is also shown. (D) Methylation analysis of the FANCF CpG island in primary ovarian carcinomas. Methylation-sensitive PCR was performed in the indicated cell lines as described in the Materials and Methods. Methylation is readily detectable in cases T27 and T43. Control unmethylated (CU) and methylated (CM) reactions are also shown.