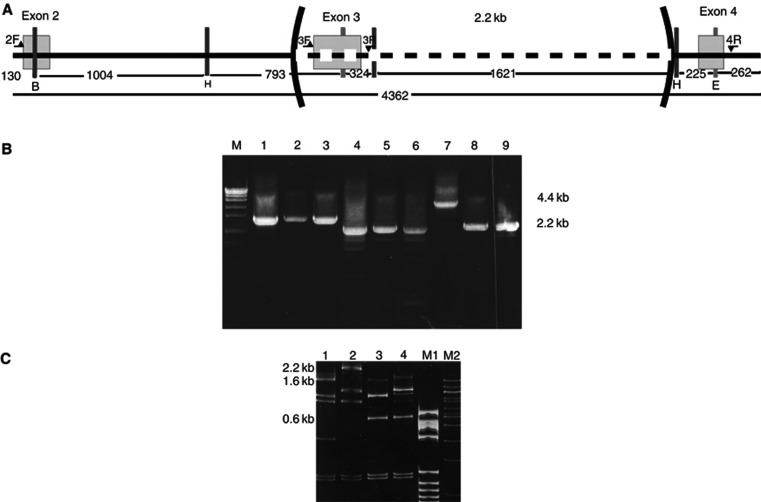
Figure 4.
Characterisation of the 2.2 kb deletion identified in family 7562. (A) Schematic representation (not to scale) of the fragment of hMSH2 gene amplified by long PCR showing the position of primers (horizontal arrows), exons (empty boxes), restriction endonuclease cleavage sites (vertical bars: H=HindIII, E=EcoRI, B=BglII) and the deletion. (B) Long-range PCR using combinations of primers (from A) spanning the repeat. M=molecular weight marker (VII, Boehringer Manheim, Germany), lanes 1–3 using primers 2F and 3R, 1=normal DNA, 2–3=patient DNA; lanes 4–6 using primers 3F and 4R, 4=normal DNA, 5–6=patient DNA; lanes 7–9 using primers 2F and 4R, 7=normal DNA, 8–9=patient DNA. (C) Restriction endonuclease digestion of PCR products from B with HindIII, EcoRI and BglII. The bands containing exon 3 (2.3 kb in lane 1 and 1.6 kb in lane 2) have disappeared and a new band 0.6 kb in length resistant to digestion with any of the three enzymes appears in the mutant sample (lanes 3 and 4). 1: normal control digested with EcoRI/HindIII/BglII. 2: normal control digested with EcoRI/HindIII. 3: patient DNA digested with EcoRI/HindIII/BglII. 4: patient DNA digested with EcoRI/HindIII. Lane M1: molecular weight marker V (Boehringer Manheim, Germany). Lane M2: Molecular weight marker 100 bp (New England Biolabs).