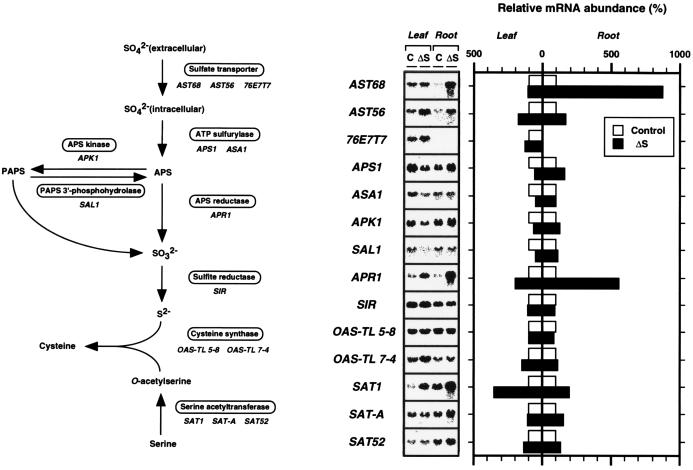
Figure 4.
Northern blot analysis of total RNA of A. thaliana. The name of the enzyme involved in each biosynthetic step and the corresponding A. thaliana cDNA clones used in this experiment are noted. Total RNA was extracted from leaves and roots of the plants grown under a control condition (C) and a sulfate-starved condition (ΔS). Three-week-old A. thaliana plants grown on GM-agar medium (MS salts, 1% sucrose; ref. 8) were transferred to sulfate-deficient GM-agar medium (MS salts as chloride instead of sulfate, 1% sucrose), which was subsequently grown for 2 days. Total RNA (20 μg) was separated in a 1.0% agarose gel containing formaldehyde, transferred to nylon membranes (Hybond N+, Amersham), and hybridized with the 32P-labeled cDNA inserts. Washing of the membranes was carried out under the same conditions as described in Fig. 4. Relative mRNA abundance was quantified by the hybridization intensities of the blots using the BAS-2000 image analyzer (Fuji). Transcript levels changed by sulfate starvation are shown in comparison with those in the control condition (100%) of leaves and roots, respectively.