Figure 3.
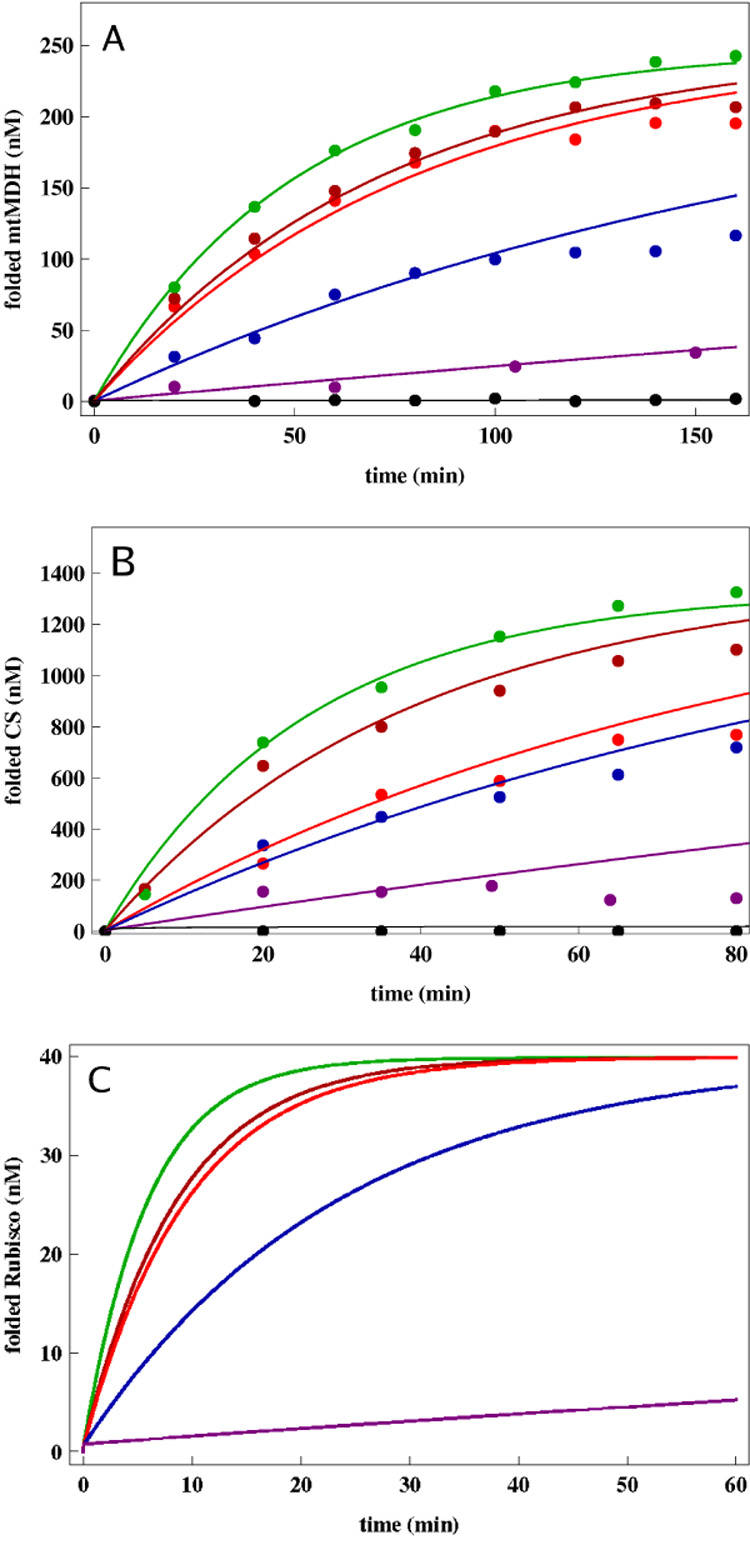
Yield of SPs as a function of time. (A) The points are taken from the experimental measurements [18] for folding mitochondrial malate dehydrogenase (mtMDH). The lines are fits to the data using the kinetic model. Black line is for spontaneous folding. Assisted folding in the presence of GroES and SR1 (purple), SR-T522I (blue) and SR-A399T (red) and SR-D115N (dark red) were used to assess the efficiencies of these three single ring chaperonins relative to GroEL (green). (B) The same as (A) except the folding of CS instead of mtMDH is analyzed. The only GroEL allosteric rate that is varied in obtaining the results in (A) and (B) is kR″−T while all others are taken from Table 1. (C) Predictions for the time-dependent yield of Rubisco for GroEL (green) and the single-ring mutants SR-T522I (blue), SR-A399T (red), SR-D115N (dark red), and SR1 (purple) based on chaperonin allosteric rates. The values of kR″−T were taken by analyzing the mtMDH data in (A). In (A) and (B) superstoichiometric concentrations of chaperonins were used.
