Abstract
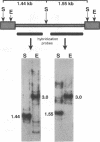
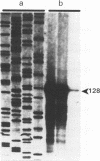
The complete sequence of ART-CH, a recently found chicken retrotransposon (A. V. Gudkov, E. A. Komarova, M. A. Nikiforov, and T. E. Zaitsevskaya, J. Virol. 66:1726-1736, 1992), was characterized. ART-CH has the structure of a 3,300-bp-long provirus, including two 388-bp long terminal repeats (LTRs) (U3, 245 bp; R region, 17 bp; and U5, 126 bp), a tRNA(Trp)-binding site, and a polypurine tract, similar to avian leukosis viruses. At least some of the approximately 50 genomic copies of ART-CH are transcribed into polyadenylated RNA, which is initiated and terminated at the expected sites within the LTRs. In contrast to the regulatory sequences involved in proviral expression and replication, the internal regions of ART-CH seem to be completely defective. Several short regions of homology with avian leukosis virus genes, most of which encode gag-related sequences, were found among different reading frames of ART-CH, which are not organized like regular retroviral genes. Both sequence analysis and restriction fragment length polymorphism analysis revealed a high degree of sequence (97% homology) and structural similarity among members of the ART-CH family, indicating their common origin and recent penetration into chicken DNA. ART-CH sequences were detected in mouse cells infected with Rous sarcoma virus produced by an ART-CH-expressing Rous sarcoma. These data are consistent with the hypothesis that ART-CH belongs to a class of defective retrotransposons whose replication strategy requires the use of helper viruses. They might originate from an avian leukosis virus-related retrovirus which completely lost its coding capacities as a result of multiple mutations and deletions. These features apparently group ART-CH with the VL30 retrotransposons of rodents.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams S. E., Rathjen P. D., Stanway C. A., Fulton S. M., Malim M. H., Wilson W., Ogden J., King L., Kingsman S. M., Kingsman A. J. Complete nucleotide sequence of a mouse VL30 retro-element. Mol Cell Biol. 1988 Aug;8(8):2989–2998. doi: 10.1128/mcb.8.8.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astrin S. M. Endogenous viral genes of the White Leghorn chicken: common site of residence and sites associated with specific phenotypes of viral gene expression. Proc Natl Acad Sci U S A. 1978 Dec;75(12):5941–5945. doi: 10.1073/pnas.75.12.5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behringer R. R., Dewey M. J. Cellular site and mode of Fv-2 gene action. II. Conditional protection of Fv-2ss cells by admixture with Fv-2rr cells. Exp Hematol. 1989 May;17(4):330–334. [PubMed] [Google Scholar]
- Boyce-Jacino M. T., O'Donoghue K., Faras A. J. Multiple complex families of endogenous retroviruses are highly conserved in the genus Gallus. J Virol. 1992 Aug;66(8):4919–4929. doi: 10.1128/jvi.66.8.4919-4929.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce-Jacino M. T., Resnick R., Faras A. J. Structural and functional characterization of the unusually short long terminal repeats and their adjacent regions of a novel endogenous avian retrovirus. Virology. 1989 Nov;173(1):157–166. doi: 10.1016/0042-6822(89)90231-6. [DOI] [PubMed] [Google Scholar]
- Brown D. W., Robinson H. L. Influence of env and long terminal repeat sequences on the tissue tropism of avian leukosis viruses. J Virol. 1988 Dec;62(12):4828–4831. doi: 10.1128/jvi.62.12.4828-4831.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buller R. S., Sitbon M., Portis J. L. The endogenous mink cell focus-forming (MCF) gp70 linked to the Rmcf gene restricts MCF virus replication in vivo and provides partial resistance to erythroleukemia induced by Friend murine leukemia virus. J Exp Med. 1988 May 1;167(5):1535–1546. doi: 10.1084/jem.167.5.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney M. G., Elder P. K., Steffen D. L., Getz M. J. Evidence for an early evolutionary origin and locus polymorphism of mouse VL30 DNA sequences. J Virol. 1982 Aug;43(2):511–518. doi: 10.1128/jvi.43.2.511-518.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney M. G., Schmidt L. J., Getz M. J. Organization and expression of endogenous virus-like (VL30) DNA sequences in nontransformed and chemically transformed mouse embryo cells in culture. Cancer Res. 1982 Feb;42(2):569–576. [PubMed] [Google Scholar]
- Dunwiddie C. T., Resnick R., Boyce-Jacino M., Alegre J. N., Faras A. J. Molecular cloning and characterization of gag-, pol-, and env-related gene sequences in the ev- chicken. J Virol. 1986 Sep;59(3):669–675. doi: 10.1128/jvi.59.3.669-675.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis R. W., Defeo D., Shih T. Y., Gonda M. A., Young H. A., Tsuchida N., Lowy D. R., Scolnick E. M. The p21 src genes of Harvey and Kirsten sarcoma viruses originate from divergent members of a family of normal vertebrate genes. Nature. 1981 Aug 6;292(5823):506–511. doi: 10.1038/292506a0. [DOI] [PubMed] [Google Scholar]
- Giri C. P., Hodgson C. P., Elder P. K., Courtney M. G., Getz M. J. Discrete regions of sequence homology between cloned rodent VL30 genetic elements and AKV-related MuLV provirus genomes. Nucleic Acids Res. 1983 Jan 25;11(2):305–319. doi: 10.1093/nar/11.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovkina T. V., Chervonsky A., Dudley J. P., Ross S. R. Transgenic mouse mammary tumor virus superantigen expression prevents viral infection. Cell. 1992 May 15;69(4):637–645. doi: 10.1016/0092-8674(92)90227-4. [DOI] [PubMed] [Google Scholar]
- Gudkov A. V., Komarova E. A., Nikiforov M. A., Zaitsevskaya T. E. ART-CH, a new chicken retroviruslike element. J Virol. 1992 Mar;66(3):1726–1736. doi: 10.1128/jvi.66.3.1726-1736.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudkov A. V., Obukh I. B., Serov S. M., Naroditsky B. S. Variety of endogenous proviruses in the genomes of chickens of different breeds. J Gen Virol. 1981 Nov;57(Pt 1):85–94. doi: 10.1099/0022-1317-57-1-85. [DOI] [PubMed] [Google Scholar]
- Hauber J., Nelböck-Hochstetter P., Feldmann H. Nucleotide sequence and characteristics of a Ty element from yeast. Nucleic Acids Res. 1985 Apr 25;13(8):2745–2758. doi: 10.1093/nar/13.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes S. H., Toyoshima K., Bishop J. M., Varmus H. E. Organization of the endogenous proviruses of chickens: implications for origin and expression. Virology. 1981 Jan 15;108(1):189–207. doi: 10.1016/0042-6822(81)90538-9. [DOI] [PubMed] [Google Scholar]
- Humphries E. H., Danhof M. L., Hlozanek I. Characterization of endogenous viral loci in five lines of white Leghorn chickens. Virology. 1984 May;135(1):125–138. doi: 10.1016/0042-6822(84)90123-5. [DOI] [PubMed] [Google Scholar]
- Ikeda H., Sugimura H. Fv-4 resistance gene: a truncated endogenous murine leukemia virus with ecotropic interference properties. J Virol. 1989 Dec;63(12):5405–5412. doi: 10.1128/jvi.63.12.5405-5412.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshet E., Itin A. Patterns of genomic distribution and sequence heterogeneity of a murine "retrovirus-like" multigene family. J Virol. 1982 Jul;43(1):50–58. doi: 10.1128/jvi.43.1.50-58.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount S. M., Rubin G. M. Complete nucleotide sequence of the Drosophila transposable element copia: homology between copia and retroviral proteins. Mol Cell Biol. 1985 Jul;5(7):1630–1638. doi: 10.1128/mcb.5.7.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick R. M., Boyce-Jacino M. T., Fu Q., Faras A. J. Phylogenetic distribution of the novel avian endogenous provirus family EAV-0. J Virol. 1990 Oct;64(10):4640–4653. doi: 10.1128/jvi.64.10.4640-4653.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovigatti U. G., Astrin S. M. Avian endogenous viral genes. Curr Top Microbiol Immunol. 1983;103:1–21. doi: 10.1007/978-3-642-68943-7_1. [DOI] [PubMed] [Google Scholar]
- Scolnick E. M., Vass W. C., Howk R. S., Duesberg P. H. Defective retrovirus-like 30S RNA species of rat and mouse cells are infectious if packaged by type C helper virus. J Virol. 1979 Mar;29(3):964–972. doi: 10.1128/jvi.29.3.964-972.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P., Gridley T., Jaenisch R. Retroviruses and insertional mutagenesis in mice: proviral integration at the Mov 34 locus leads to early embryonic death. Genes Dev. 1987 Jun;1(4):366–375. doi: 10.1101/gad.1.4.366. [DOI] [PubMed] [Google Scholar]
- Spence S. E., Gilbert D. J., Swing D. A., Copeland N. G., Jenkins N. A. Spontaneous germ line virus infection and retroviral insertional mutagenesis in eighteen transgenic Srev lines of mice. Mol Cell Biol. 1989 Jan;9(1):177–184. doi: 10.1128/mcb.9.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoye J. P., Coffin J. M. The four classes of endogenous murine leukemia virus: structural relationships and potential for recombination. J Virol. 1987 Sep;61(9):2659–2669. doi: 10.1128/jvi.61.9.2659-2669.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoye J. P., Fenner S., Greenoak G. E., Moran C., Coffin J. M. Role of endogenous retroviruses as mutagens: the hairless mutation of mice. Cell. 1988 Jul 29;54(3):383–391. doi: 10.1016/0092-8674(88)90201-2. [DOI] [PubMed] [Google Scholar]
- Tereba A. Asymmetric chromosomal distribution of endogenous retrovirus loci in chickens and mice. Curr Top Microbiol Immunol. 1983;107:29–50. doi: 10.1007/978-3-642-69075-4_2. [DOI] [PubMed] [Google Scholar]
- Tsichlis P. N., Lazo P. A. Virus-host interactions and the pathogenesis of murine and human oncogenic retroviruses. Curr Top Microbiol Immunol. 1991;171:95–171. doi: 10.1007/978-3-642-76524-7_5. [DOI] [PubMed] [Google Scholar]
- Yuki S., Inouye S., Ishimaru S., Saigo K. Nucleotide sequence characterization of a Drosophila retrotransposon, 412. Eur J Biochem. 1986 Jul 15;158(2):403–410. doi: 10.1111/j.1432-1033.1986.tb09767.x. [DOI] [PubMed] [Google Scholar]