Figure 5.
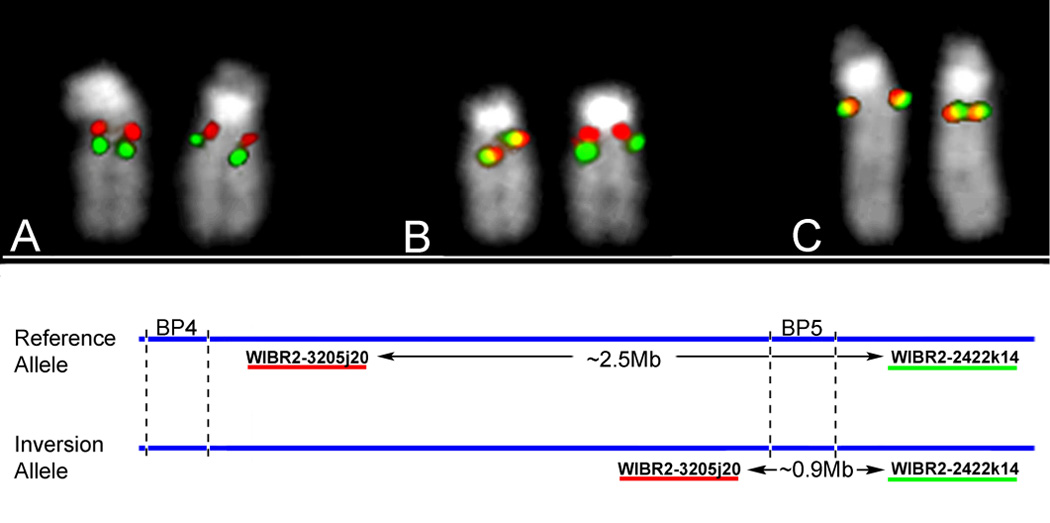
Identification of a common inversion polymorphism in 15q13.3. To detect inversions of the BP4-BP5 microdeletion region, we performed FISH mapping using fosmid probes located proximal and distal to BP5. The separation of these probes in the reference assembly is ~2.5 Mb, enough to visualize these as two separate signals on metaphase chromosomes. Inversion of the region BP4-BP5 moves the two probes within close proximity to each other, visualized as overlapping yellow signals. Using this assay on 8 HapMap individuals of different ethnicities, we observed the inversion on 7/16 chromosomes (~44% of the population, Supplementary Table 2). We also tested the mother of patient 69/06 (in whose germline the BP4-BP5 deletion arose), who was found to be heterozygous. Shown are individuals (a) homozygous for the reference allele, (b) heterozygous, or (c) homozygous for the inversion allele.
