Fig. 3. Characterization of the SMIT TonEs.
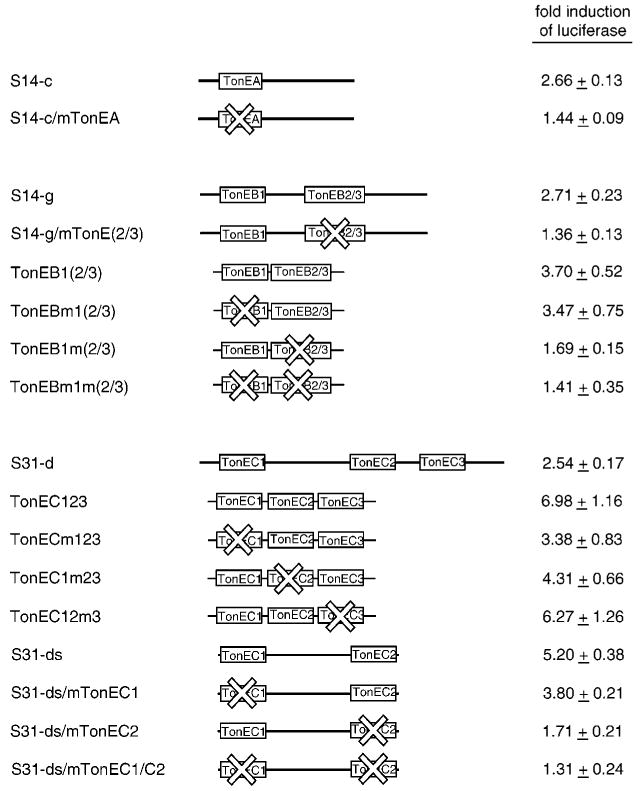
DNA fragments S14-c (top), S14-g (middle), and S31-d (bottom) are shown as thick lines with positions of TonE sequences indicated (not to scale) as boxes. S31-ds was derived from S31-d using PCR. TonEA in S14-c and TonEB(2/3) in S14-g were inactivated (as indicated by X marks) by mutating key nucleotide residues as described under “Experimental Procedures” to generate S14-c/mTonEA and S14-g/mTonE(2/3), respectively. Likewise, TonEC1 and TonEC2 in S31-ds were mutated to generate S31-ds/mTonEC1, S31-ds/mTonEC2, and S31-ds/mTonEC1/C2. TonEB1(2/3) (middle) and TonEC123 (bottom) are described in Table I. In other DNAs, TonE sequences marked by X were mutated by changing the 2nd G to T, the 5th A to T, and the 9th pyrimidine to G. Each of the wild-type and mutant DNA fragments was tested for its tonicity-responsive enhancer activity as described in Fig. 2. -Fold induction of luciferase in response to hypertonicity in cells transfected with each construct is shown: mean ± S.E., n = 4–9.
