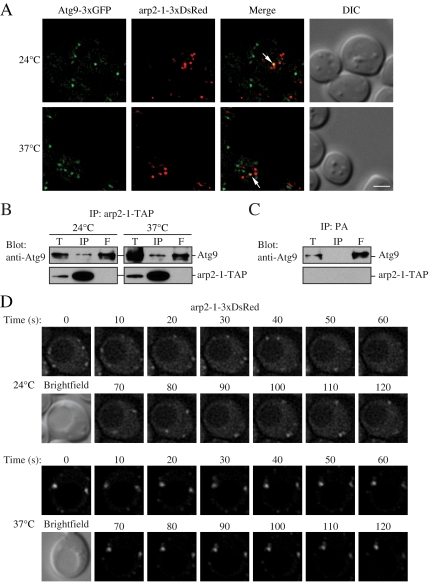
Figure 7.
Atg9 interacts with arp2-1, which is defective in movement. (A) Atg9-3xGFP and arp2-1-3xDsRed colocalize. arp2-1 cells expressing chromosomally tagged Atg9-3xGFP and arp2-1-3xDsRed (JGY086) were grown to mid-log phase at 24°C, and then they were shifted to 37°C for 30 min. Samples were collected before and after the temperature shift, and then they were analyzed by microscopy as described in Materials and Methods. Colocalization of Atg9-3xGFP and arp2-1-3xDsRed in a single Z-section is indicated by arrows. (B) The arp2-1 protein interacts with Atg9. Cells (IRA038) expressing the chromosomally TAP-tagged arp2-1 protein were grown in SMD medium at 24°C, and then one aliquot of the culture was shifted to 37°C for 3 h. Cells were collected, and affinity isolation was performed as in Materials and Methods. The same amounts of total lysate (T), immunoprecipitate (IP), and flow through (F) were separated by SDS-PAGE and detected by anti-Atg9 antiserum. Bar, 2 μm. (C) Atg9 is not coimmunoprecipitated with protein A (PA) alone. Wild-type cells expressing CUP1 promoter-driven PA were used as a negative control for Atg9 interaction. Cells were cultured at 30°C and analyzed by affinity isolation as described in B. (D) arp2-1-DsRed is defective in movement at the nonpermissive temperature. Mid-log phase cells expressing arp2-1-3xDsRed (JGY086) were grown and imaged as in described in A with a time interval between Z-stack acquisitions of 10 s for 120-s duration. Still frames from collapsed series Z-sections examine arp2-1-3xDsRed movement at 24 and 37°C. Bar, 2 μm.