Figure 3.
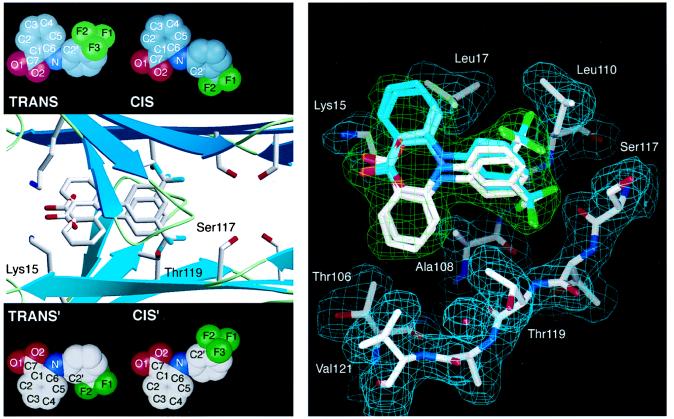
(Left) Flu–TTR interactions. Ribbon diagram of the tetrameric TTR with Flu showing a closeup of one of the two funnel-shaped binding cavities computed by the program setor (32). The binding site is formed by two adjacent TTR monomers related by a twofold axis of symmetry (from left to right in the paper plane). Residues Ser-117, Thr-119, and Lys-15, which are displaced on binding of Flu, are highlighted. For clarity, only the trans conformation of Flu is shown with its symmetry mate. Insets show all four observed binding conformations (the molecular conformations cis and trans with the symmetry related binding modes cis′ and trans′). (Right) TTR conformational changes mediated by Flu binding. A stick model of the Flu binding site as computed by o (33). Only residues from one subunit of TTR that align the binding cavity are displayed, with their 2 |Fo| − |Fc| Φcalc density in cyan. Flu is shown in all four of its binding modes. Because of the twofold-symmetry axis (from left to right in paper plane) both molecular conformations of Flu fit in two symmetry-related binding modes into the final 2 |Fo| − |Fc| Φcalc in green. Oxygen atoms are red, nitrogen atoms are blue, fluorine atoms are green, and carbon atoms are shown in white and cyan, respectively. The protein–ligand interactions are described in the text.
